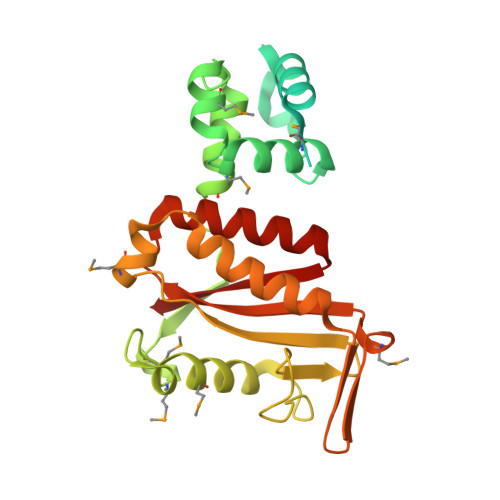
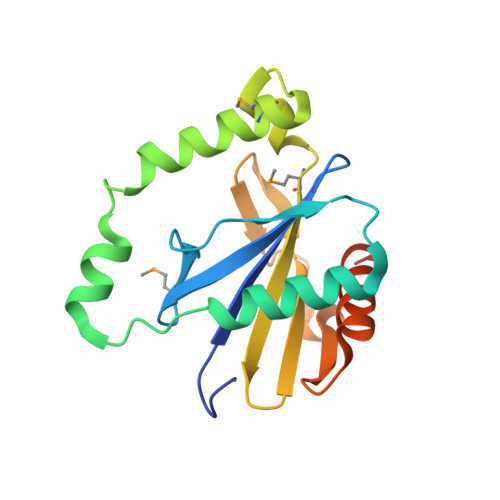
Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems.
Morse, R.P., Nikolakakis, K.C., Willett, J.L., Gerrick, E., Low, D.A., Hayes, C.S., Goulding, C.W.(2012) Proc Natl Acad Sci U S A 109: 21480-21485
- PubMed: 23236156
- DOI: https://doi.org/10.1073/pnas.1216238110
- Primary Citation of Related Structures:
4G6U, 4G6V - PubMed Abstract:
Contact-dependent growth inhibition (CDI) systems encode polymorphic toxin/immunity proteins that mediate competition between neighboring bacterial cells. We present crystal structures of CDI toxin/immunity complexes from Escherichia coli EC869 and Burkholderia pseudomallei 1026b. Despite sharing little sequence identity, the toxin domains are structurally similar and have homology to endonucleases. The EC869 toxin is a Zn(2+)-dependent DNase capable of completely degrading the genomes of target cells, whereas the Bp1026b toxin cleaves the aminoacyl acceptor stems of tRNA molecules. Each immunity protein binds and inactivates its cognate toxin in a unique manner. The EC869 toxin/immunity complex is stabilized through an unusual β-augmentation interaction. In contrast, the Bp1026b immunity protein exploits shape and charge complementarity to occlude the toxin active site. These structures represent the initial glimpse into the CDI toxin/immunity network, illustrating how sequence-diverse toxins adopt convergent folds yet retain distinct binding interactions with cognate immunity proteins. Moreover, we present visual demonstration of CDI toxin delivery into a target cell.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697, USA.