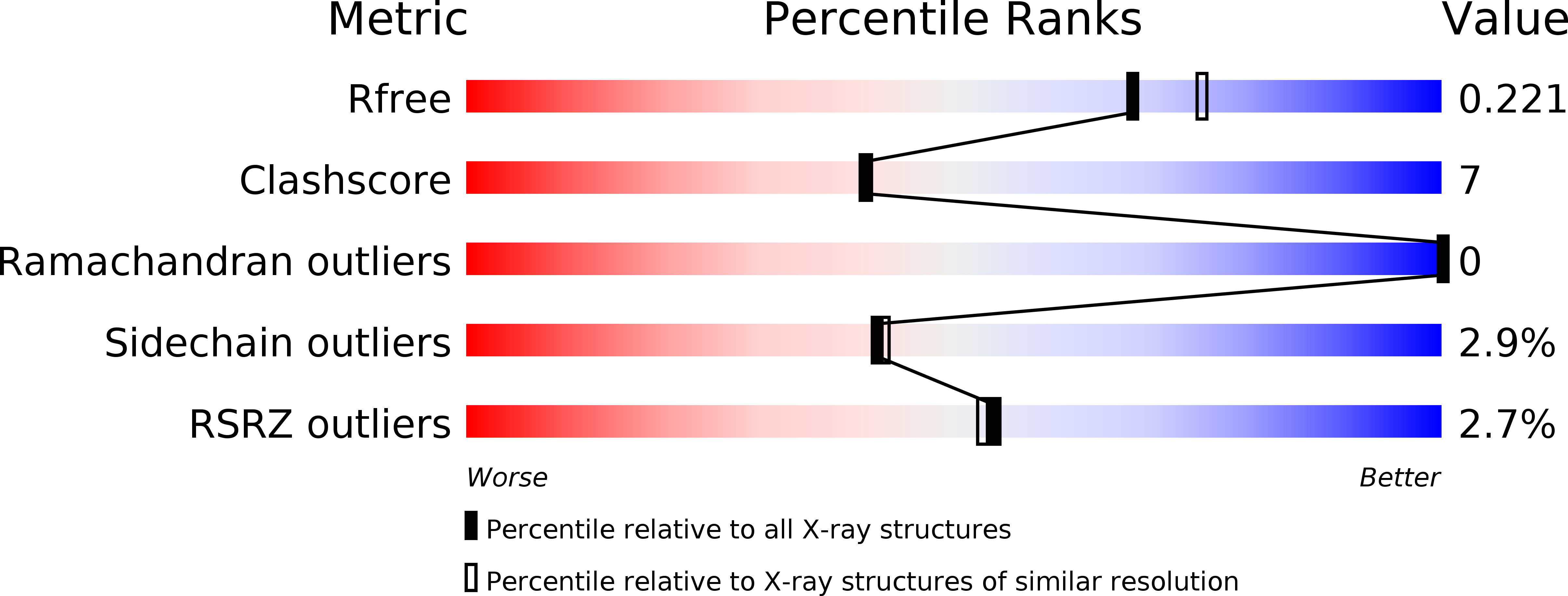
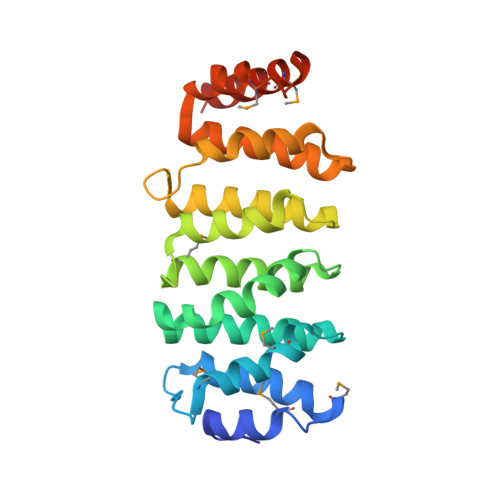
The structure of the TOG-like domain of Drosophila melanogaster Mast/Orbit.
De la Mora-Rey, T., Guenther, B.D., Finzel, B.C.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 723-729
- PubMed: 23832196
- DOI: https://doi.org/10.1107/S1744309113015182
- Primary Citation of Related Structures:
4G3A - PubMed Abstract:
Mast/Orbit is a nonmotor microtubule-associated protein (MAP) present in Drosophila melanogaster that reportedly binds microtubules at the plus end and is essential for mitosis. Sequence analysis has shown that the N-terminal domain (Mast-M1) resembles TOG domains from the Dis1-TOG family of proteins and stands as a representative of one of the three subclasses of divergent TOG-like domains (TOGL1) that includes human CLASP1. The crystal structure of Mast-M1 has been determined at 2.0 Å resolution and provides the first detailed structural description of any TOG-like domain. The structure confirms that Mast-M1 adopts a similar fold to the previously described Dis1-TOG domains of microtubule-binding proteins. A comparison with three known TOG-domain structures from XMAP215/Dis1 family members exposes significant differences between Mast-M1 and other TOG-domain structures in key residues at the proposed tubulin-binding edge.
Organizational Affiliation:
Department of Medicinal Chemistry, University of Minnesota, 308 Harvard Street SE, 8-101 Weaver-Densford Hall, Minneapolis, MN 55455, USA.