Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Elshahawi, S.I., Ramelot, T.A., Seetharaman, J., Chen, J., Singh, S., Yang, Y., Pederson, K., Kharel, M.K., Xiao, R., Lew, S., Yennamalli, R.M., Miller, M.D., Wang, F., Tong, L., Montelione, G.T., Kennedy, M.A., Bingman, C.A., Zhu, H., Phillips, G.N., Thorson, J.S.(2014) ACS Chem Biol 9: 2347-2358
- PubMed: 25079510
- DOI: https://doi.org/10.1021/cb500327m
- Primary Citation of Related Structures:
2LUZ, 4FPW - PubMed Abstract:
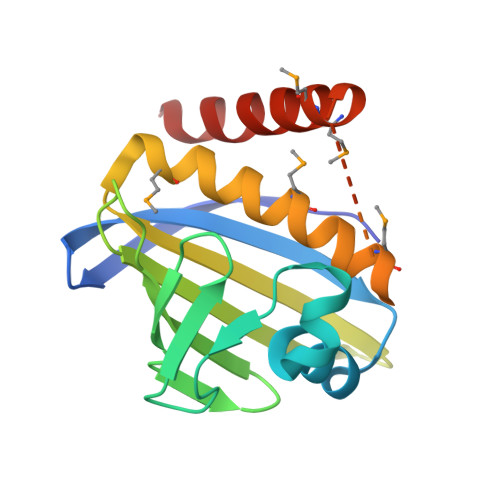
Calicheamicin γ1I (1) is an enediyne antitumor compound produced by Micromonospora echinospora spp. calichensis, and its biosynthetic gene cluster has been previously reported. Despite extensive analysis and biochemical study, several genes in the biosynthetic gene cluster of 1 remain functionally unassigned. Using a structural genomics approach and biochemical characterization, two proteins encoded by genes from the 1 biosynthetic gene cluster assigned as "unknowns", CalU16 and CalU19, were characterized. Structure analysis revealed that they possess the STeroidogenic Acute Regulatory protein related lipid Transfer (START) domain known mainly to bind and transport lipids and previously identified as the structural signature of the enediyne self-resistance protein CalC. Subsequent study revealed calU16 and calU19 to confer resistance to 1, and reminiscent of the prototype CalC, both CalU16 and CalU19 were cleaved by 1 in vitro. Through site-directed mutagenesis and mass spectrometry, we identified the site of cleavage in each protein and characterized their function in conferring resistance against 1. This report emphasizes the importance of structural genomics as a powerful tool for the functional annotation of unknown proteins.
Organizational Affiliation:
Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky , Lexington, Kentucky 40536, United States.