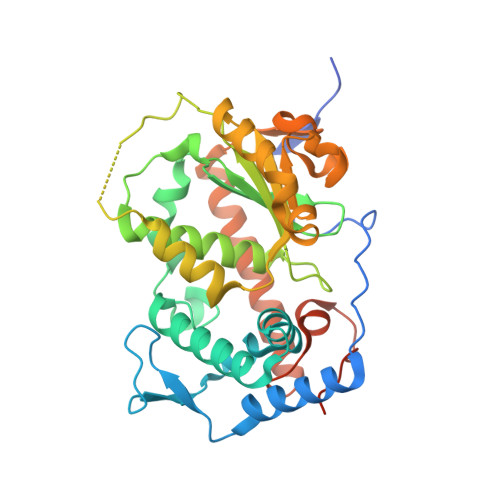
Dimeric Sfh3 has structural changes in its binding pocket that are associated with a dimer-monomer state transformation induced by substrate binding.
Yuan, Y., Zhao, W., Wang, X., Gao, Y., Niu, L., Teng, M.(2013) Acta Crystallogr D Biol Crystallogr 69: 313-323
- PubMed: 23519406
- DOI: https://doi.org/10.1107/S0907444912046161
- Primary Citation of Related Structures:
4FMM - PubMed Abstract:
Phosphorylated derivatives of phosphatidylinositol (PtdIns), also called phosphoinositides (PIPs), are basic components of membrane-associated signalling systems. A family of PtdIns-transfer proteins (PITPs) called the Sec14 family have been predicted to form a set of functional modules that can sense different types of lipid metabolism and transmit the information to the PIP signalling system. In eukaryotic cells, the Sec14 family exhibits a wide diversity of activity, but the structural basis of this diversity remains unclear. In the present study, the dimeric structure of Sfh3 (Sec14 family homologue 3 in yeast) is reported for the first time and differs from the Sec14 proteins reported to date, all of which are monomeric. Some variations in the binding pocket of Sfh3 were observed and the dimer interface was identified and proposed to provide a link between dimer-monomer state changes and PtdIns binding. Together, these structural changes and the oligomeric state transformation of Sfh3 support ideas of diversity within the Sec14 family and provide some new clues to function.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, People's Republic of China.