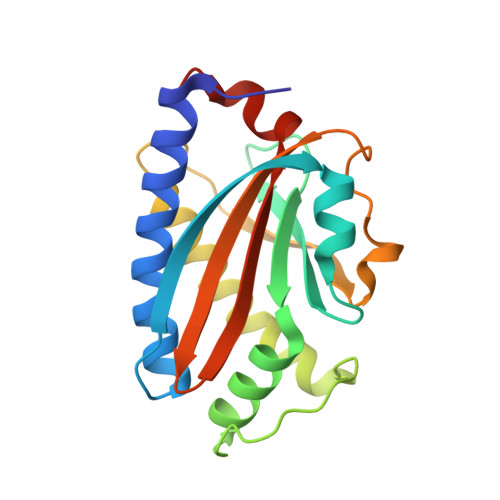
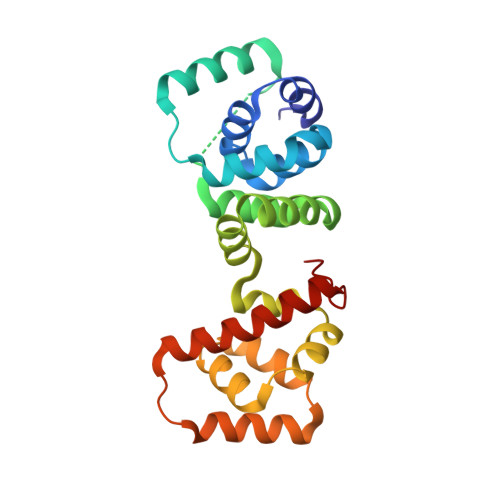
Structure of flagellar motor proteins in complex allows for insights into motor structure and switching.
Vartanian, A.S., Paz, A., Fortgang, E.A., Abramson, J., Dahlquist, F.W.(2012) J Biol Chem 287: 35779-35783
- PubMed: 22896702
- DOI: https://doi.org/10.1074/jbc.C112.378380
- Primary Citation of Related Structures:
4FHR - PubMed Abstract:
The flagellar motor is one type of propulsion device of motile bacteria. The cytoplasmic ring (C-ring) of the motor interacts with the stator to generate torque in clockwise and counterclockwise directions. The C-ring is composed of three proteins, FliM, FliN, and FliG. Together they form the "switch complex" and regulate switching and torque generation. Here we report the crystal structure of the middle domain of FliM in complex with the middle and C-terminal domains of FliG that shows the interaction surface and orientations of the proteins. In the complex, FliG assumes a compact conformation in which the middle and C-terminal domains (FliG(MC)) collapse and stack together similarly to the recently published structure of a mutant of FliG(MC) with a clockwise rotational bias. This intramolecular stacking of the domains is distinct from the intermolecular stacking seen in other structures of FliG. We fit the complex structure into the three-dimensional reconstructions of the motor and propose that the cytoplasmic ring is assembled from 34 FliG and FliM molecules in a 1:1 fashion.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Barbara, California 93106, USA.