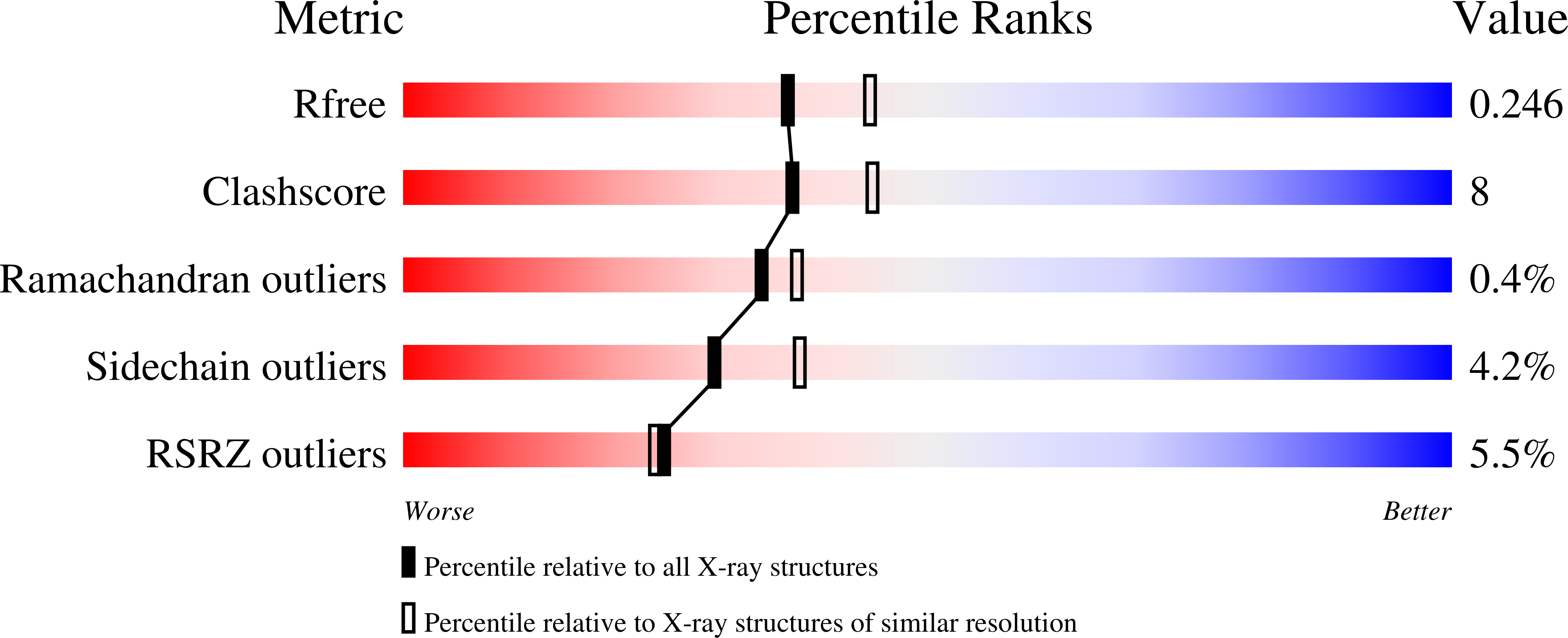
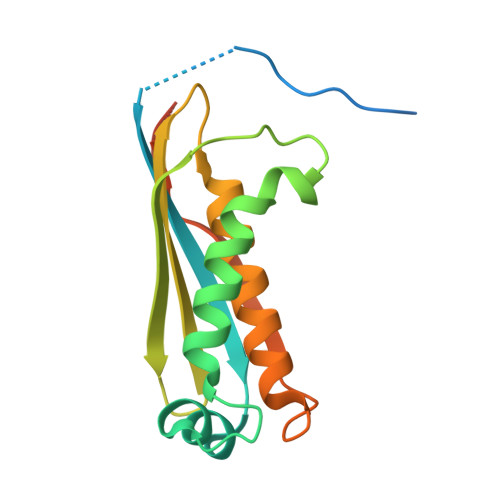
Structural Insights into Putative Molybdenum Cofactor Biosynthesis Protein C (MoaC2) from Mycobacterium tuberculosis H37Rv.
Srivastava, V.K., Srivastava, S., Arora, A., Pratap, J.V.(2013) PLoS One 8: e58333-e58333
- PubMed: 23526978
- DOI: https://doi.org/10.1371/journal.pone.0058333
- Primary Citation of Related Structures:
4FDF - PubMed Abstract:
The Molybdenum cofactor (Moco) biosynthesis pathway is an evolutionary conserved pathway seen in almost all eukaryotes including the pathogenic species Mycobacterium tuberculosis. This pathway comprises of several novel reactions which include the initial formation of precursor Z from guanosine triphosphate (GTP), catalysed by two enzymes MoaA and MoaC. Although Moco biosynthesis is well understood, the first step is still not clear. In M. tuberculosis H37Rv, three orthologous genes of MoaC have been annotated: moaC1 (Rv3111), moaC2 (Rv0864) and moaC3 (Rv3324c). Rv0864 (MoaC2) is a 17.5 kDa protein and is reported to be down-regulated by ∼3 times in the nutrient starvation model for Mycobacterium tuberculosis. The crystal structure of Moco-biosynthesis protein MoaC2 from Mycobacterium tuberculosis (2.20 Å resolution, space group P213) has been determined. Based on a comparative analysis of structures of homologous proteins, conserved residues were identified and are implicated in structural and functional roles. Molecular docking studies with probable ligands carried out in order to identify its ligand, suggests that pteridinebenzomonophosphate as the most likely ligand. Sequence based interaction study identified MoaA1 to interact with MoaC2. A homology model of MoaA1 was then complexed with MoaC2 and protein-protein interactions are also discussed.
Organizational Affiliation:
Molecular and Structural Biology Division, CSIR-Central Drug Research Institute, Mahatma Gandhi Marg, Lucknow, India.