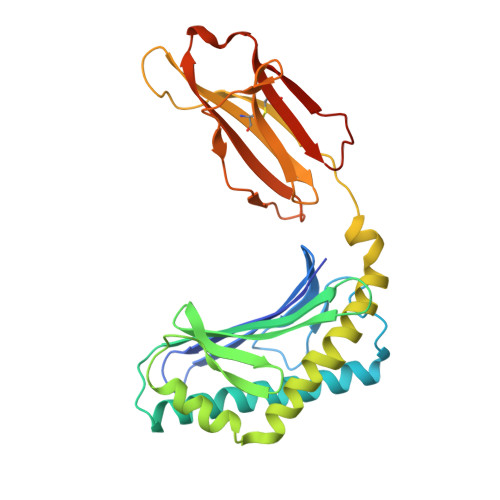
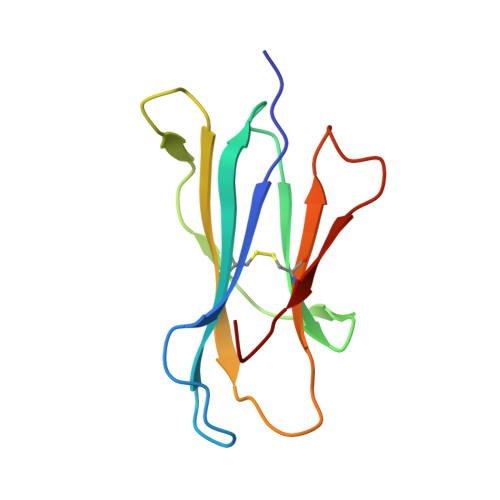
Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Wang, J., Guillaume, J., Pauwels, N., Van Calenbergh, S., Van Rhijn, I., Zajonc, D.M.(2012) PLoS One 7: e47989-e47989
- PubMed: 23110152
- DOI: https://doi.org/10.1371/journal.pone.0047989
- Primary Citation of Related Structures:
4F7C, 4F7E - PubMed Abstract:
NKT cells play important roles in immune surveillance. They rapidly respond to pathogens by detecting microbial glycolipids when presented by the non-classical MHC I homolog CD1d. Previously, ruminants were considered to lack NKT cells due to the lack of a functional CD1D gene. However, recent data suggest that cattle express CD1d with unknown function. In an attempt to characterize the function of bovine CD1d, we assessed the lipid binding properties of recombinant Bos taurus CD1d (boCD1d) in vitro. BoCD1d is able to bind glycosphingolipids (GSLs) with fatty acid chain lengths of C₁₈, while GSLs with fatty acids of C₂₄ do not bind. Crystal structures of boCD1d bound to a short-chain C₁₂-di-sulfatide antigen, as well as short-chain C₁₆-αGalCer revealed that the Á pocket of boCD1d is restricted in size compared to that of both mouse and human CD1d, explaining the inability of long chain GSL's to bind to boCD1d. Moreover, while di-sulfatide is presented similarly compared to the presentation of sulfatide by mouse CD1d, αGalCer is presented differently at the cell surface, due to an amino acid Asp151Asn substitution that results in loss of intimate contacts between the αGalCer headgroup and CD1d. The altered αGalCer presentation by boCD1d also explains its lack of cross-activation of mouse iNKT cells and raises the interesting question of the nature and function of bovine lipid-reactive T cells.
Organizational Affiliation:
Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, California, United States of America.