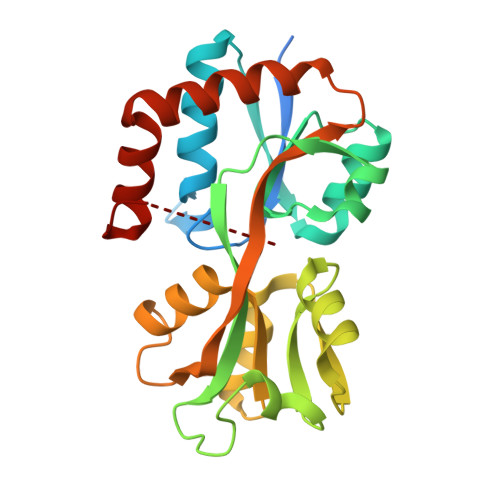
Crystal structure of a Glutamine-binding periplasmic protein from Burkholderia pseudomallei in complex with glutamine
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J., Sankaran, B., Fairman, J.W., Staker, B., Myler, P., Stewart, L.To be published.