Crystal Structure of the Largest Subunit of a Bacterial RNA-guided Immune Complex and Its Role in DNA Target Binding.
Mulepati, S., Orr, A., Bailey, S.(2012) J Biol Chem 287: 22445-22449
- PubMed: 22621933
- DOI: https://doi.org/10.1074/jbc.C112.379503
- Primary Citation of Related Structures:
4F3E - PubMed Abstract:
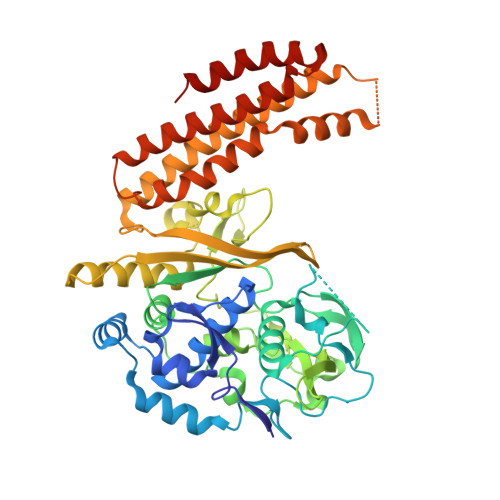
Prokaryotes make use of small RNAs encoded by CRISPR (clustered regularly interspaced short palindromic repeat) loci to provide immunity against bacteriophage or plasmid invasion. In Escherichia coli, the CRISPR-associated complex for antiviral defense (Cascade) utilizes these RNAs to target foreign DNA for destruction. CasA, the largest subunit of Cascade, is essential for its function. Here we report the crystal structure of Thermus thermophilus CasA. The structure is composed of two domains that are arranged in a chair-like conformation with a novel fold forming the larger N-terminal domain. Docking of the crystal structure into cryo-electron microscopy maps reveals two loops in CasA that likely have important functions in DNA target binding. Finally, DNA binding experiments show that CasA is essential for binding of Cascade to DNA target.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Johns Hopkins School of Public Health, Baltimore, Maryland 21205, USA.