The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium
Wu, R., Jedrzejczak, R.P., Brown, R.N., Cort, J.R., Heffron, F., Nakayasu, E.S., Adkins, J.N., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Conjugative transfer: regulation | 127 | Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | Mutation(s): 0 Gene Names: STM14_5595, traJ | 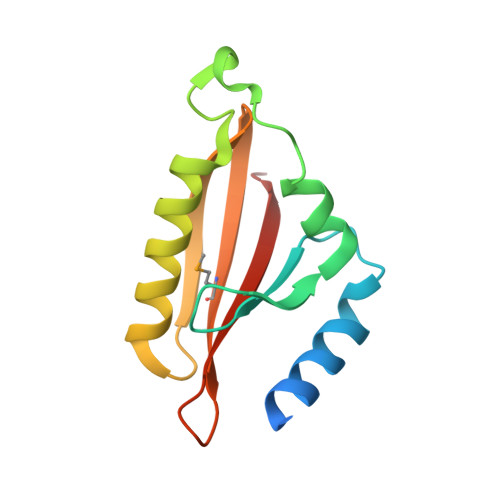 | |
UniProt | |||||
Find proteins for A0A0F6AW83 (Salmonella typhimurium (strain 14028s / SGSC 2262)) Explore A0A0F6AW83 Go to UniProtKB: A0A0F6AW83 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0F6AW83 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SIN Query on SIN | B [auth A] | SUCCINIC ACID C4 H6 O4 KDYFGRWQOYBRFD-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | G [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| PYR Query on PYR | D [auth A] | PYRUVIC ACID C3 H4 O3 LCTONWCANYUPML-UHFFFAOYSA-N |  | ||
| ACY Query on ACY | C [auth A] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | E [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.081 | α = 90 |
| b = 57.081 | β = 90 |
| c = 67.638 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-3000 | data collection |
| MLPHARE | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |