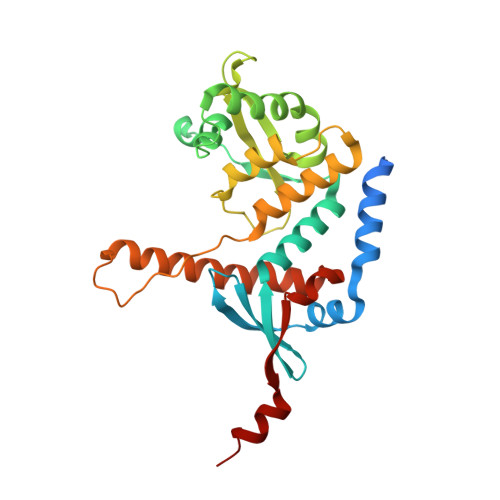
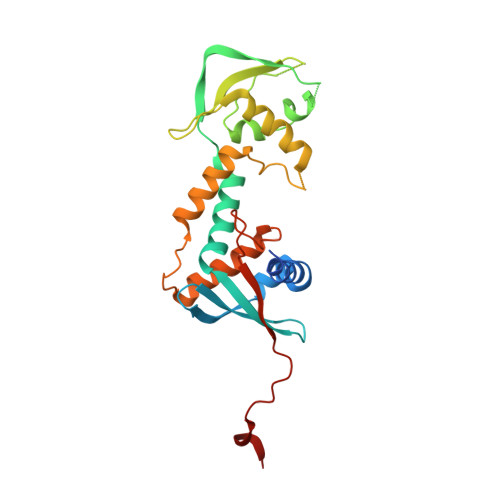
Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Gueneau, E., Dherin, C., Legrand, P., Tellier-Lebegue, C., Gilquin, B., Bonnesoeur, P., Londino, F., Quemener, C., Le Du, M.H., Marquez, J.A., Moutiez, M., Gondry, M., Boiteux, S., Charbonnier, J.B.(2013) Nat Struct Mol Biol 20: 461-468
- PubMed: 23435383
- DOI: https://doi.org/10.1038/nsmb.2511
- Primary Citation of Related Structures:
4E4W, 4FMN, 4FMO - PubMed Abstract:
Mismatch-repair factors have a prominent role in surveying eukaryotic DNA-replication fidelity and in ensuring correct meiotic recombination. These functions depend on MutL-homolog heterodimers with Mlh1. In humans, MLH1 mutations underlie half of hereditary nonpolyposis colorectal cancers (HNPCCs). Here we report crystal structures of the MutLα (Mlh1-Pms1 heterodimer) C-terminal domain (CTD) from Saccharomyces cerevisiae, alone and in complex with fragments derived from Mlh1 partners. These structures reveal structural rearrangements and additional domains in MutLα as compared to the bacterial MutL counterparts and show that the strictly conserved C terminus of Mlh1 forms part of the Pms1 endonuclease site. The structures of the ternary complexes between MutLα(CTD) and Exo1 or Ntg2 fragments reveal the binding mode of the MIP-box motif shared by several Mlh1 partners. Finally, the structures provide a rationale for the deleterious impact of MLH1 mutations in HNPCCs.
Organizational Affiliation:
Unité Mixte de Recherche 8221, Commissariat à l'Energie Atomique, Centre National de la Recherche Scientifique, Institut de Biologie et Technologies de Saclay, Gif-sur-Yvette, France.