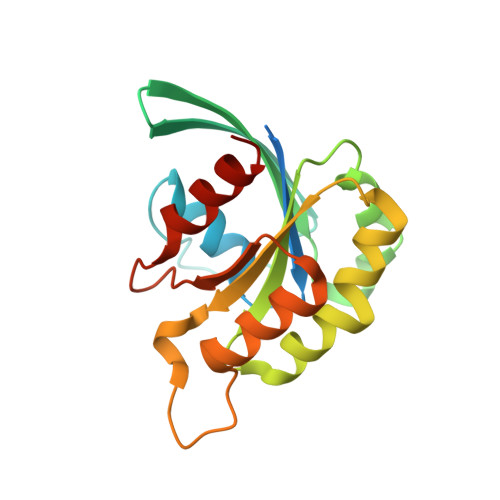
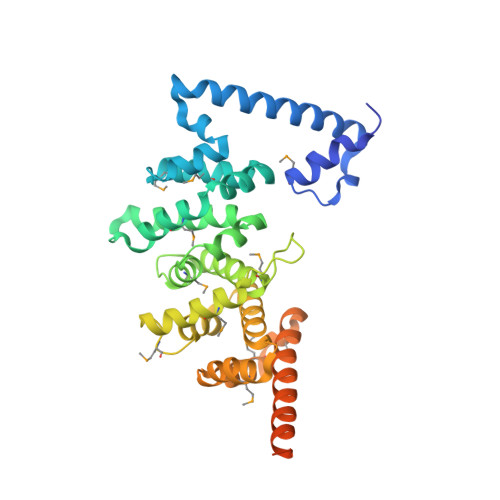
Entamoeba histolytica Rho1 Regulates Actin Polymerization through a Divergent, Diaphanous-Related Formin.
Bosch, D.E., Yang, B., Siderovski, D.P.(2012) Biochemistry 51: 8791-8801
- PubMed: 23050667
- DOI: https://doi.org/10.1021/bi300954g
- Primary Citation of Related Structures:
4DVG - PubMed Abstract:
Entamoeba histolytica requires a dynamic actin cytoskeleton for intestinal and systemic pathogenicity. Diaphanous-related formins represent an important family of actin regulators that are activated by Rho GTPases. The E. histolytica genome encodes a large family of Rho GTPases and three diaphanous-related formins, of which EhFormin1 is known to regulate mitosis and cytokinesis in trophozoites. We demonstrate that EhFormin1 modulates actin polymerization through its formin homology 2 domain. Despite a highly divergent diaphanous autoinhibitory domain, EhFormin1 is autoinhibited by an N- and C-terminal intramolecular interaction but activated upon binding of EhRho1 to the N-terminal domain tandem. A crystal structure of the EhRho1·GTPγS-EhFormin1 complex illustrates an EhFormin1 conformation that diverges from mammalian mDia1 and lacks a secondary interaction with a Rho insert helix. The structural model also highlights residues required for specific recognition of the EhRho1 GTPase and suggests that the molecular mechanisms of EhFormin1 autoinhibition and activation differ from those of mammalian homologues.
Organizational Affiliation:
Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599-7365, United States.