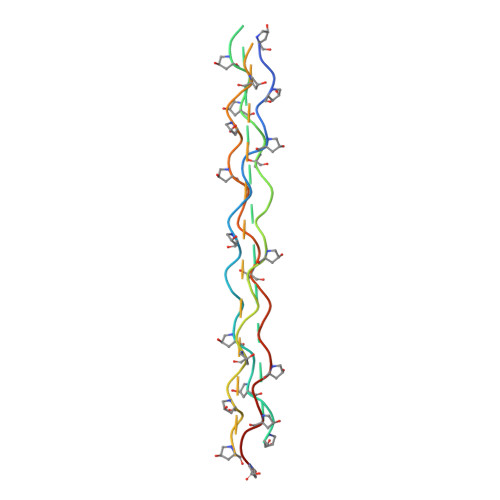
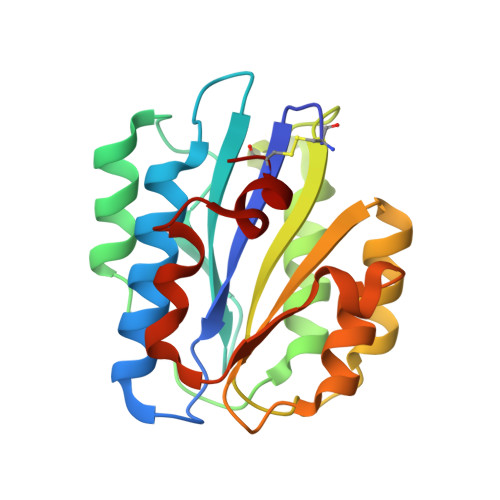
Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex.
Brondijk, T.H., Bihan, D., Farndale, R.W., Huizinga, E.G.(2012) Proc Natl Acad Sci U S A 109: 5253-5258
- PubMed: 22440751
- DOI: https://doi.org/10.1073/pnas.1112388109
- Primary Citation of Related Structures:
4DMT, 4DMU - PubMed Abstract:
Fibrillar collagens, the most abundant proteins in the vertebrate body, are involved in a plethora of biological interactions. Plasma protein von Willebrand factor (VWF) mediates adhesion of blood platelets to fibrillar collagen types I, II, and III, which is essential for normal haemostasis. High affinity VWF-binding sequences have been identified in the homotrimeric collagen types II and III, however, it is unclear how VWF recognizes the heterotrimeric collagen type I, the superstructure of which is unknown. Here we present the crystal structure of VWF domain A3 bound to a collagen type III-derived homotrimeric peptide. Our structure reveals that VWF-A3 interacts with all three collagen chains and binds through conformational selection to a sequence that is one triplet longer than was previously appreciated from platelet and VWF binding studies. The VWF-binding site overlaps those of SPARC (also known as osteonectin) and discodin domain receptor 2, but is more extended and shifted toward the collagen amino terminus. The observed collagen-binding mode of VWF-A3 provides direct structural constraints on collagen I chain registry. A VWF-binding site can be generated from the sequences RGQAGVMF, present in the two α1(I) chains, and RGEOGNIGF, in the unique α2(I) chain, provided that α2(I) is in the middle or trailing position. Combining these data with previous structural data on integrin binding to collagen yields strong support for the trailing position of the α2(I) chain, shedding light on the fundamental and long-standing question of the collagen I chain registry.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH, Utrecht, The Netherlands.