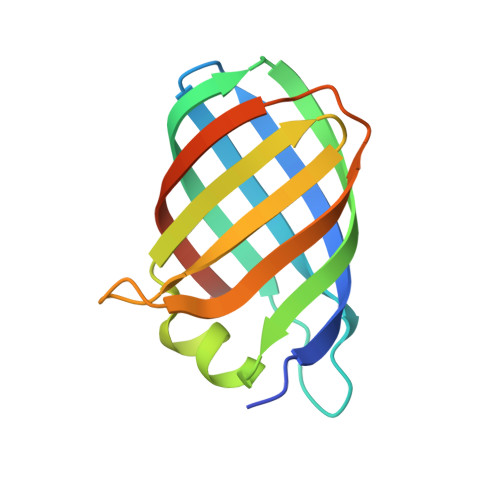
Structure of the Virulence-Associated Protein Vapd from the Intracellular Pathogen Rhodococcus Equi.
Whittingham, J.L., Blagova, E.V., Finn, C.E., Luo, H., Miranda-Casoluengo, R., Turkenburg, J.P., Leech, A.P., Walton, P.H., Murzin, A.G., Meijer, W.G., Wilkinson, A.J.(2014) Acta Crystallogr D Biol Crystallogr 70: 2139
- PubMed: 25084333
- DOI: https://doi.org/10.1107/S1399004714012632
- Primary Citation of Related Structures:
4CSB - PubMed Abstract:
Rhodococcus equi is a multi-host pathogen that infects a range of animals as well as immune-compromised humans. Equine and porcine isolates harbour a virulence plasmid encoding a homologous family of virulence-associated proteins associated with the capacity of R. equi to divert the normal processes of endosomal maturation, enabling bacterial survival and proliferation in alveolar macrophages. To provide a basis for probing the function of the Vap proteins in virulence, the crystal structure of VapD was determined. VapD is a monomer as determined by multi-angle laser light scattering. The structure reveals an elliptical, compact eight-stranded β-barrel with a novel strand topology and pseudo-twofold symmetry, suggesting evolution from an ancestral dimer. Surface-associated octyl-β-D-glucoside molecules may provide clues to function. Circular-dichroism spectroscopic analysis suggests that the β-barrel structure is preceded by a natively disordered region at the N-terminus. Sequence comparisons indicate that the core folds of the other plasmid-encoded virulence-associated proteins from R. equi strains are similar to that of VapD. It is further shown that sequences encoding putative R. equi Vap-like proteins occur in diverse bacterial species. Finally, the functional implications of the structure are discussed in the light of the unique structural features of VapD and its partial structural similarity to other β-barrel proteins.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5DD, England.