Structural and Functional Analysis of the Three Mif4G Domains of Nonsense-Mediated Decay Factor Upf2.
Clerici, M., Deniaud, A., Boehm, V., Gehring, N.H., Schaffitzel, C., Cusack, S.(2014) Nucleic Acids Res 42: 2673
- PubMed: 24271394
- DOI: https://doi.org/10.1093/nar/gkt1197
- Primary Citation of Related Structures:
4CEK, 4CEM - PubMed Abstract:
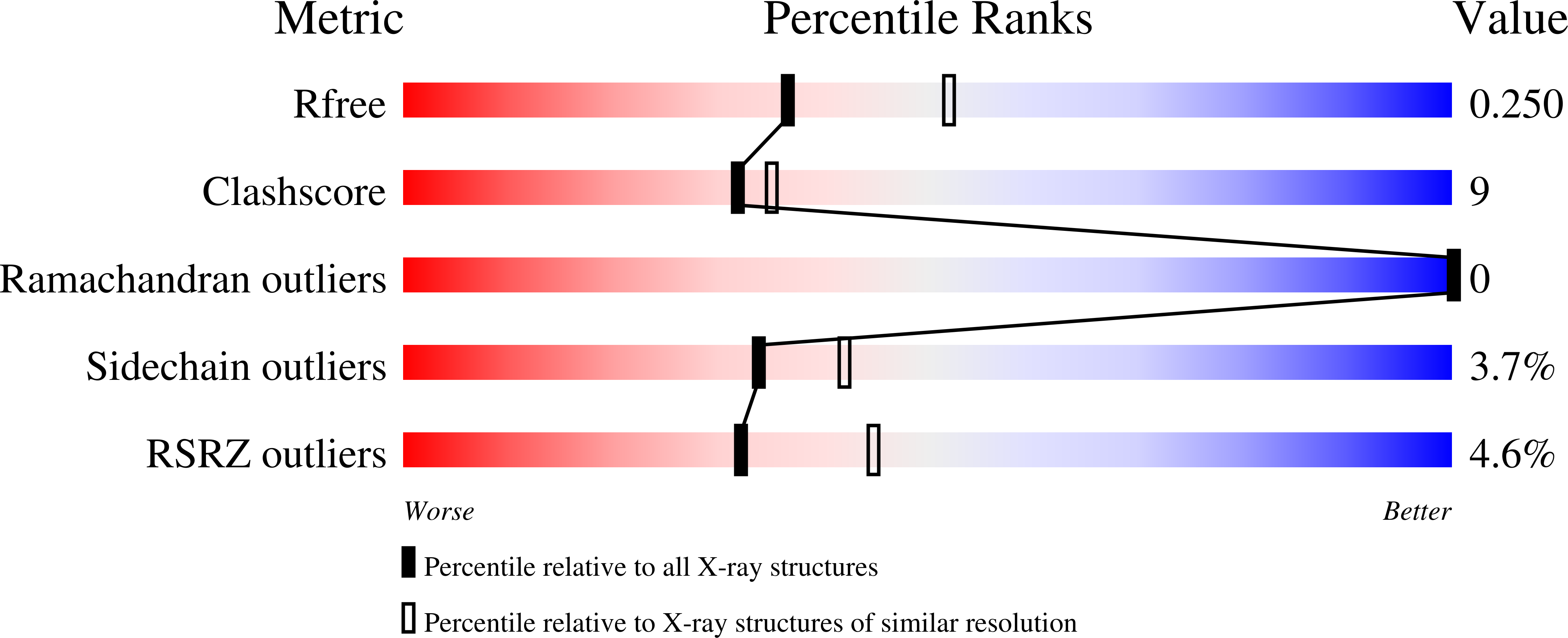
Nonsense-mediated decay (NMD) is a eukaryotic quality control pathway, involving conserved proteins UPF1, UPF2 and UPF3b, which detects and degrades mRNAs with premature stop codons. Human UPF2 comprises three tandem MIF4G domains and a C-terminal UPF1 binding region. MIF4G-3 binds UPF3b, but the specific functions of MIF4G-1 and MIF4G-2 are unknown. Crystal structures show that both MIF4G-1 and MIF4G-2 contain N-terminal capping helices essential for stabilization of the 10-helix MIF4G core and that MIF4G-2 interacts with MIF4G-3, forming a rigid assembly. The UPF2/UPF3b/SMG1 complex is thought to activate the kinase SMG1 to phosphorylate UPF1 in vivo. We identify MIF4G-3 as the binding site and in vitro substrate of SMG1 kinase and show that a ternary UPF2 MIF4G-3/UPF3b/SMG1 complex can form in vitro. Whereas in vivo complementation assays show that MIF4G-1 and MIF4G-2 are essential for NMD, tethering assays reveal that UPF2 truncated to only MIF4G-3 and the UPF1-binding region can still partially accomplish NMD. Thus UPF2 MIF4G-1 and MIF4G-2 appear to have a crucial scaffolding role, while MIF4G-3 is the key module required for triggering NMD.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble Outstation, 6 rue Jules Horowitz, 38042 Grenoble Cedex 9, France, Unit of Virus Host-Cell Interactions, University of Grenoble Alpes-EMBL-CNRS, UMI 3265, 6 rue Jules Horowitz, 38042 Grenoble Cedex 9, France and University of Cologne, Institute for Genetics, Zuelpicher Street 47a, 50674 Cologne, Germany.