H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Van Der Vries, E., Collins, P.J., Vachieri, S.G., Xiong, X., Liu, J., Walker, P.A., Haire, L.F., Hay, A.J., Schutten, M., Osterhaus, A.D.M.E., Martin, S.R., Boucher, C.A.B., Skehel, J.J., Gamblin, S.J.(2012) PLoS Pathog 8: 2914
- PubMed: 23028314
- DOI: https://doi.org/10.1371/journal.ppat.1002914
- Primary Citation of Related Structures:
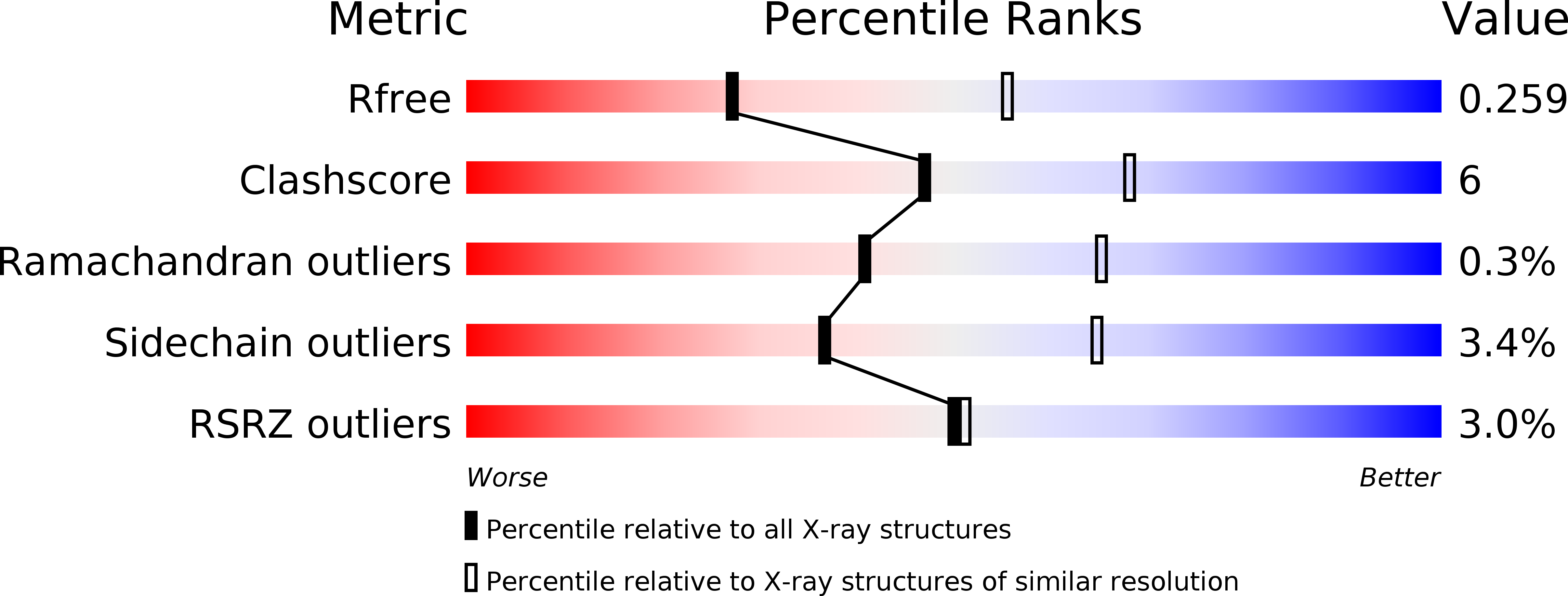
4B7J, 4B7M, 4B7N, 4B7Q, 4B7R - PubMed Abstract:
Two classes of antiviral drugs, neuraminidase inhibitors and adamantanes, are approved for prophylaxis and therapy against influenza virus infections. A major concern is that antiviral resistant viruses emerge and spread in the human population. The 2009 pandemic H1N1 virus is already resistant to adamantanes. Recently, a novel neuraminidase inhibitor resistance mutation I223R was identified in the neuraminidase of this subtype. To understand the resistance mechanism of this mutation, the enzymatic properties of the I223R mutant, together with the most frequently observed resistance mutation, H275Y, and the double mutant I223R/H275Y were compared. Relative to wild type, K(M) values for MUNANA increased only 2-fold for the single I223R mutant and up to 8-fold for the double mutant. Oseltamivir inhibition constants (K(I)) increased 48-fold in the single I223R mutant and 7500-fold in the double mutant. In both cases the change was largely accounted for by an increased dissociation rate constant for oseltamivir, but the inhibition constants for zanamivir were less increased. We have used X-ray crystallography to better understand the effect of mutation I223R on drug binding. We find that there is shrinkage of a hydrophobic pocket in the active site as a result of the I223R change. Furthermore, R223 interacts with S247 which changes the rotamer it adopts and, consequently, binding of the pentoxyl substituent of oseltamivir is not as favorable as in the wild type. However, the polar glycerol substituent present in zanamivir, which mimics the natural substrate, is accommodated in the I223R mutant structure in a similar way to wild type, thus explaining the kinetic data. Our structural data also show that, in contrast to a recently reported structure, the active site of 2009 pandemic neuraminidase can adopt an open conformation.
Organizational Affiliation:
Erasmus Medical Centre, Department of Virology, Rotterdam, The Netherlands.