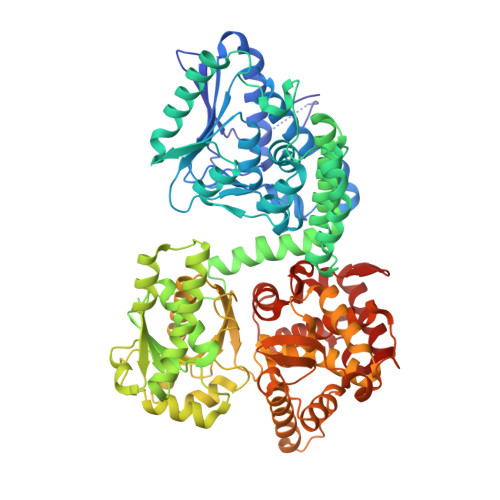
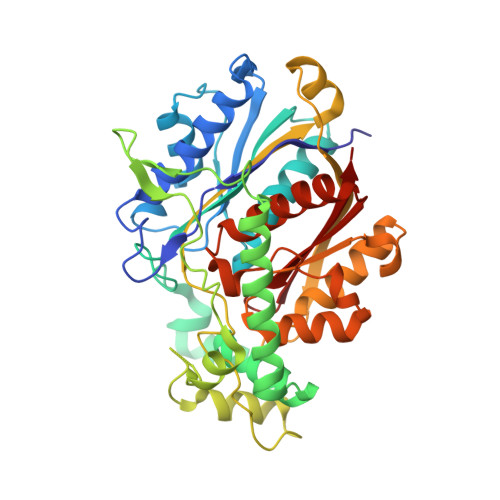
Structure of Mycobacterial Beta-Oxidation Trifunctional Enzyme Reveals its Altered Assembly and Putative Substrate Channeling Pathway.
Venkatesan, R., Wierenga, R.K.(2013) ACS Chem Biol 8: 1063
- PubMed: 23496842
- DOI: https://doi.org/10.1021/cb400007k
- Primary Citation of Related Structures:
4B3H, 4B3I, 4B3J - PubMed Abstract:
The incidence of tuberculosis is increasing due to the appearance of new drug-resistant variants. A thorough understanding of the disease organism is essential in order to create more effective drugs. In an attempt to understand better the poorly studied lipid metabolism of Mycobacterium tuberculosis (Mtb), we identified and characterized its fatty acid β-oxidation complex (trifunctional enzyme (TFE)). TFE is an α(2)β(2) complex consisting of two types of polypeptides catalyzing three of the four reactions of the β-oxidation of fatty acids. The kinetic constants (k(cat) and K(m)) show that the complexed α chain is more active than the individual α chain. Crystal structures of Mtb TFE (mtTFE) reveal that the quaternary assembly is strikingly different from the already known Pseudomonas fragi TFE (pfTFE) assembly due to the presence of a helical insertion (LA5) in the mtTFE-β subunit. This helical insertion prevents the pfTFE mode of assembly, as it would clash with helix H9A of the TFE-α chain. The mtTFE assembly appears to be more rigid and results in a different substrate channeling path between the α and the β subunits. Structural comparisons suggest that the mtTFE active sites can accommodate bulkier fatty acyl chains than in pfTFE. Although another thiolase (FadA2), more closely related to human TFE-β/thiolase, is present in the Mtb genome, it does not form a complex with mtTFE-α. Extensive phylogenetic analyses show that there are at least four TFE subfamilies. Our studies highlight the molecular properties of mtTFE, significantly extending the structural knowledge on this type of very interesting multifunctional enzymes.
Organizational Affiliation:
Department of Biochemistry and Biocenter Oulu, University of Oulu, Oulu 90014, Finland.