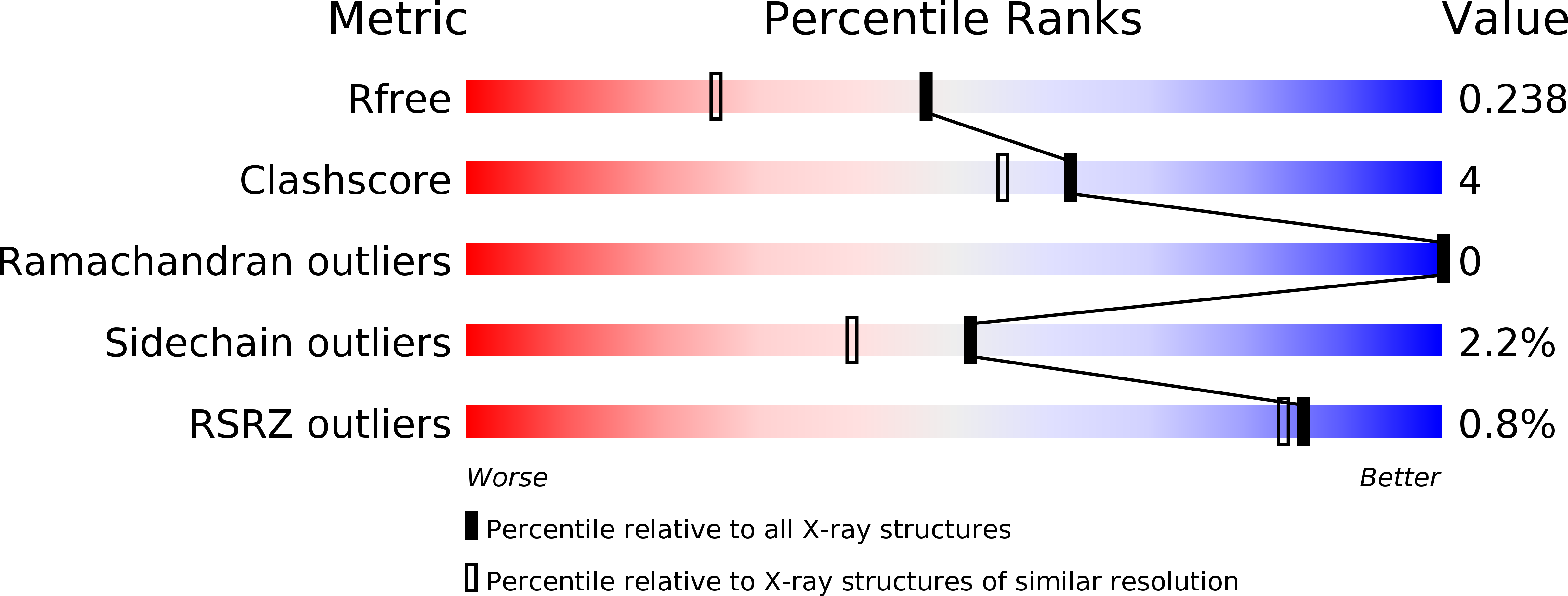
Design and Synthesis of Inhibitors of Plasmodium Falciparum N-Myristoyltransferase, a Promising Target for Anti-Malarial Drug Discovery.
Yu, Z., Brannigan, J.A., Moss, D.K., Brzozowski, A.M., Wilkinson, A.J., Holder, A.A., Tate, E.W., Leatherbarrow, R.J.(2012) J Med Chem 55: 8879
- PubMed: 23035716
- DOI: https://doi.org/10.1021/jm301160h
- Primary Citation of Related Structures:
4B10, 4B11, 4B12, 4B13, 4B14 - PubMed Abstract:
Design of inhibitors for N-myristoyltransferase (NMT), an enzyme responsible for protein trafficking in Plasmodium falciparum , the most lethal species of parasites that cause malaria, is described. Chemistry-driven optimization of compound 1 from a focused NMT inhibitor library led to the identification of two early lead compounds 4 and 25, which showed good enzyme and cellular potency and excellent selectivity over human NMT. These molecules provide a valuable starting point for further development.
Organizational Affiliation:
Department of Chemistry, Imperial College London, London SW7 2AZ, UK.