Visualizing the Substrate-, Superoxo-, Alkylperoxo- and Product-Bound States at the Non-Heme Fe(II) Site of Homogentisate Dioxygenase
Jeoung, J.-H., Bommer, M., Lin, T.-Y., Dobbek, H.(2013) Proc Natl Acad Sci U S A 110: 12625
- PubMed: 23858455
- DOI: https://doi.org/10.1073/pnas.1302144110
- Primary Citation of Related Structures:
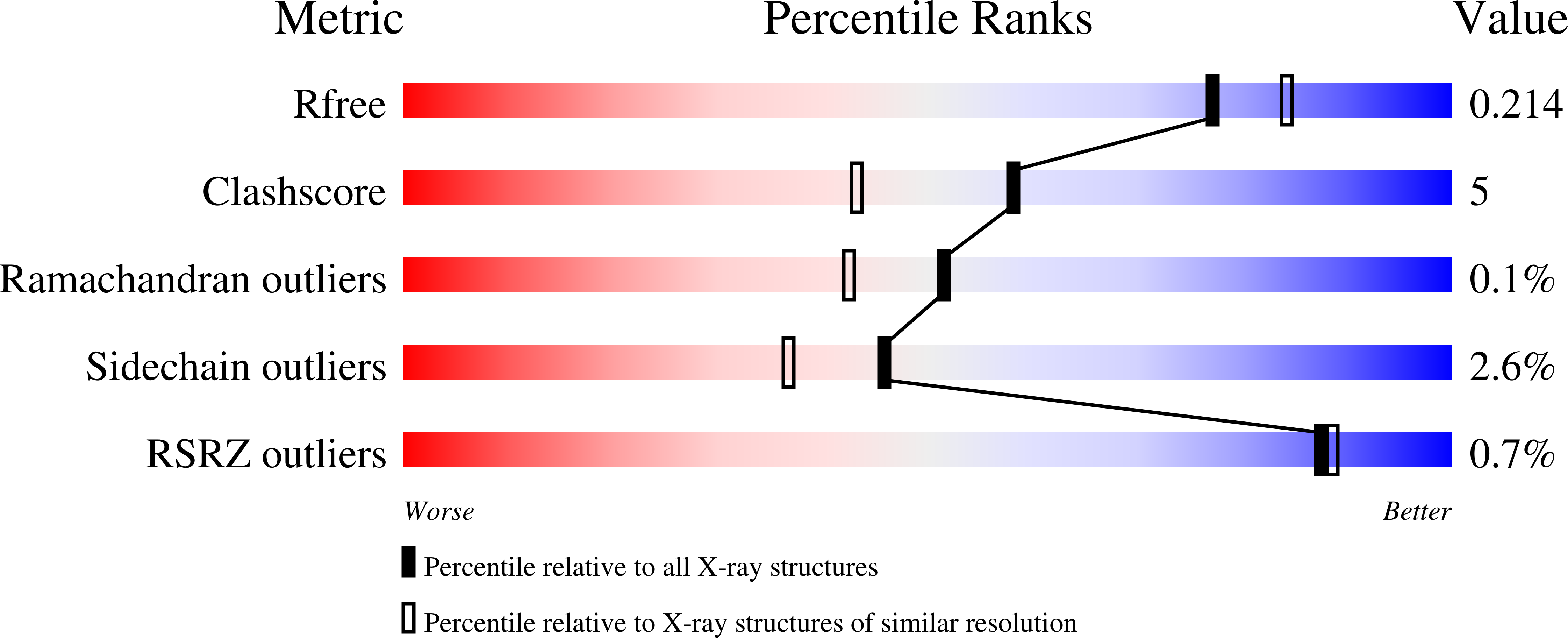
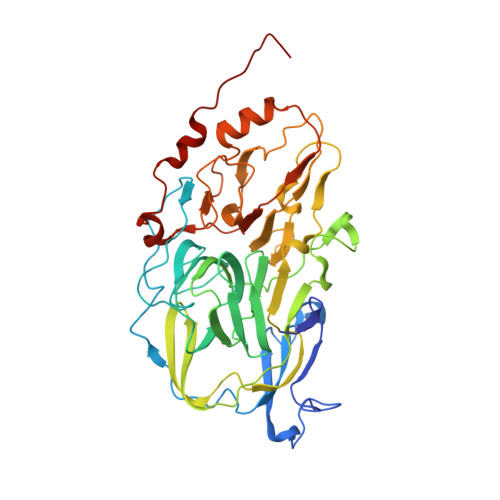
3ZDS, 4AQ2, 4AQ6 - PubMed Abstract:
Homogentisate 1,2-dioxygenase (HGDO) uses a mononuclear nonheme Fe(2+) to catalyze the oxidative ring cleavage in the degradation of Tyr and Phe by producing maleylacetoacetate from homogentisate (2,5-dihydroxyphenylacetate). Here, we report three crystal structures of HGDO, revealing five different steps in its reaction cycle at 1.7-1.98 Å resolution. The resting state structure displays an octahedral coordination for Fe(2+) with two histidine residues (His331 and His367), a bidentate carboxylate ligand (Glu337), and two water molecules. Homogentisate binds as a monodentate ligand to Fe(2+), and its interaction with Tyr346 invokes the folding of a loop over the active site, effectively shielding it from solvent. Binding of homogentisate is driven by enthalpy and is entropically disfavored as shown by anoxic isothermal titration calorimetry. Three different reaction cycle intermediates have been trapped in different HGDO subunits of a single crystal showing the influence of crystal packing interactions on the course of enzymatic reactions. The observed superoxo:semiquinone-, alkylperoxo-, and product-bound intermediates have been resolved in a crystal grown anoxically with homogentisate, which was subsequently incubated with dioxygen. We demonstrate that, despite different folds, active site architectures, and Fe(2+) coordination, extradiol dioxygenases can proceed through the same principal reaction intermediates to catalyze the O2-dependent cleavage of aromatic rings. Thus, convergent evolution of nonhomologous enzymes using the 2-His-1-carboxylate facial triad motif developed different solutions to stabilize closely related intermediates in unlike environments.
Organizational Affiliation:
Institut für Biologie, Strukturbiologie/Biochemie, Humboldt-Universität zu Berlin, 10099 Berlin, Germany.