A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Park, J.-T., Song, H.-N., Jung, T.-Y., Lee, M.H., Park, S.G., Woo, E.-J., Park, K.-H.(2013) Biochim Biophys Acta 1834: 380
- PubMed: 22902546
- DOI: https://doi.org/10.1016/j.bbapap.2012.08.001
- Primary Citation of Related Structures:
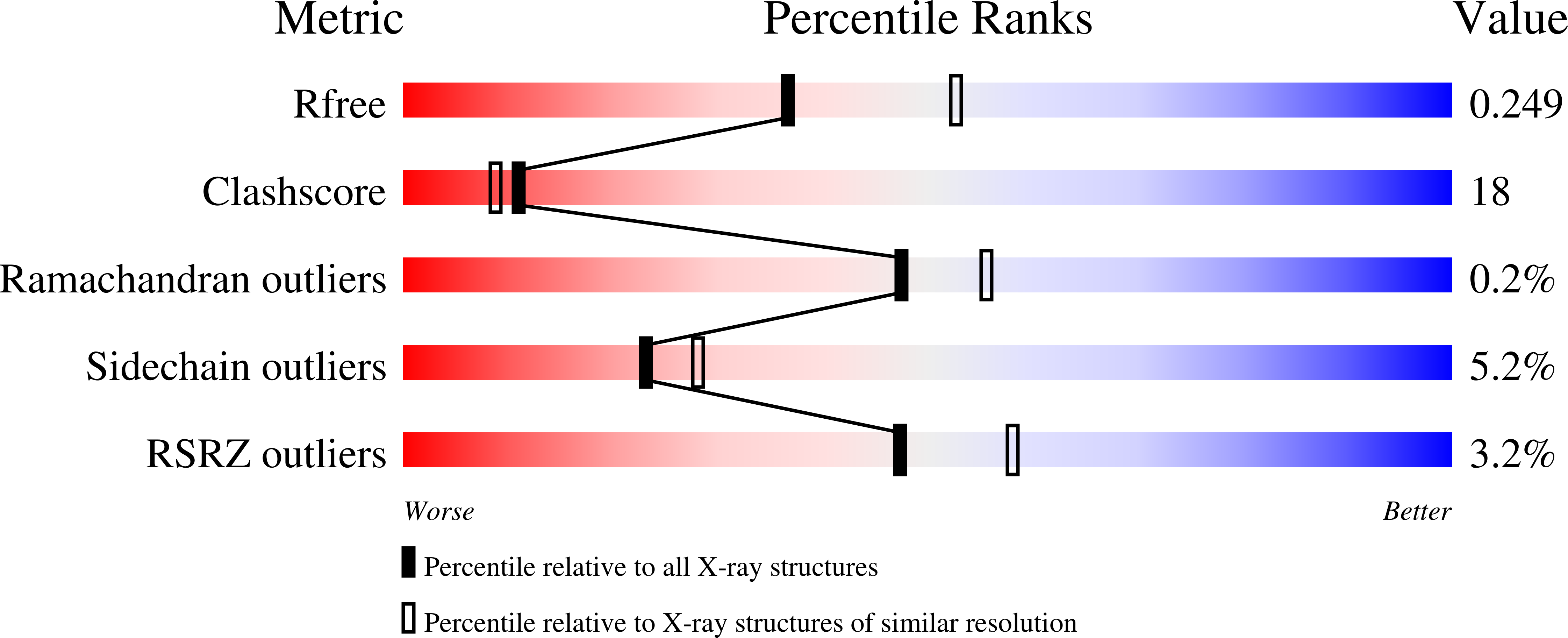
4AEF - PubMed Abstract:
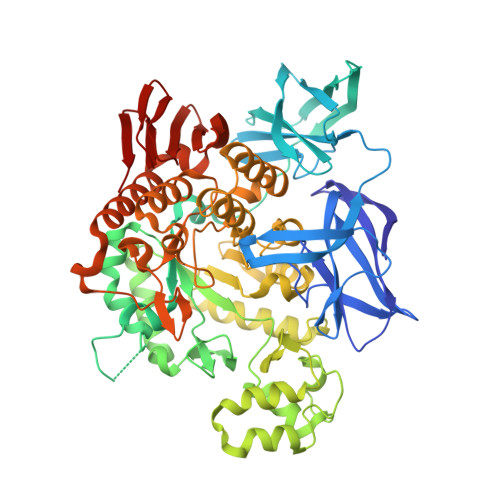
PFTA (Pyrococcus furiosus thermostable amylase) is a hyperthermophilic amylase isolated from the archaeon Pyrococcus furiosus. This enzyme possesses characteristics of both α-amylase- and cyclodextrin (CD)-hydrolyzing enzymes, allowing it to degrade pullulan, CD and acarbose-activities that are absent in most α-amylases-without the transferring activity that is common in CD-hydrolyzing enzymes. The crystal structure of PFTA revealed a unique monomeric subunit with an extended N-terminal region and an N'-domain folded into its own active site-a significantly altered domain configuration relative to that of the conventional dimeric CD-hydrolyzing amylases in glycoside hydrolase family 13. The active site is formed by the interface of the N'-domain and the catalytic domain and exhibits a broad and wide-open geometry without the concave pocket that is commonly found in the active sites of maltogenic amylases. The mutation of a residue (Gly415 to Glu) located at the domain interface between the N'- and catalytic domains yielded an enzyme that produced a significantly higher purity maltoheptaose (G7) from β-CD, supporting the involvement of this interface in substrate recognition and indicating that this mutant enzyme is a suitable candidate for the production of pure G7. The unique configuration of the active site distinguishes this archaic monomeric enzyme from classical bacterial CD-hydrolyzing amylases and provides a molecular basis for its enzymatic characteristics and for its potential use in industrial applications.
Organizational Affiliation:
Department of Food Science and Technology, Chungnam National University, 99 Daehak-ro, Yuseong-gu, Daejeon 305-764, Republic of Korea.