Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Guo, X., Peisker, K., Backbro, K., Chen, Y., Kiran, R.K., Mandava, C.S., Sanyal, S., Selmer, M.(2012) Open Biol 2: 20016
- PubMed: 22645663
- DOI: https://doi.org/10.1098/rsob.120016
- Primary Citation of Related Structures:
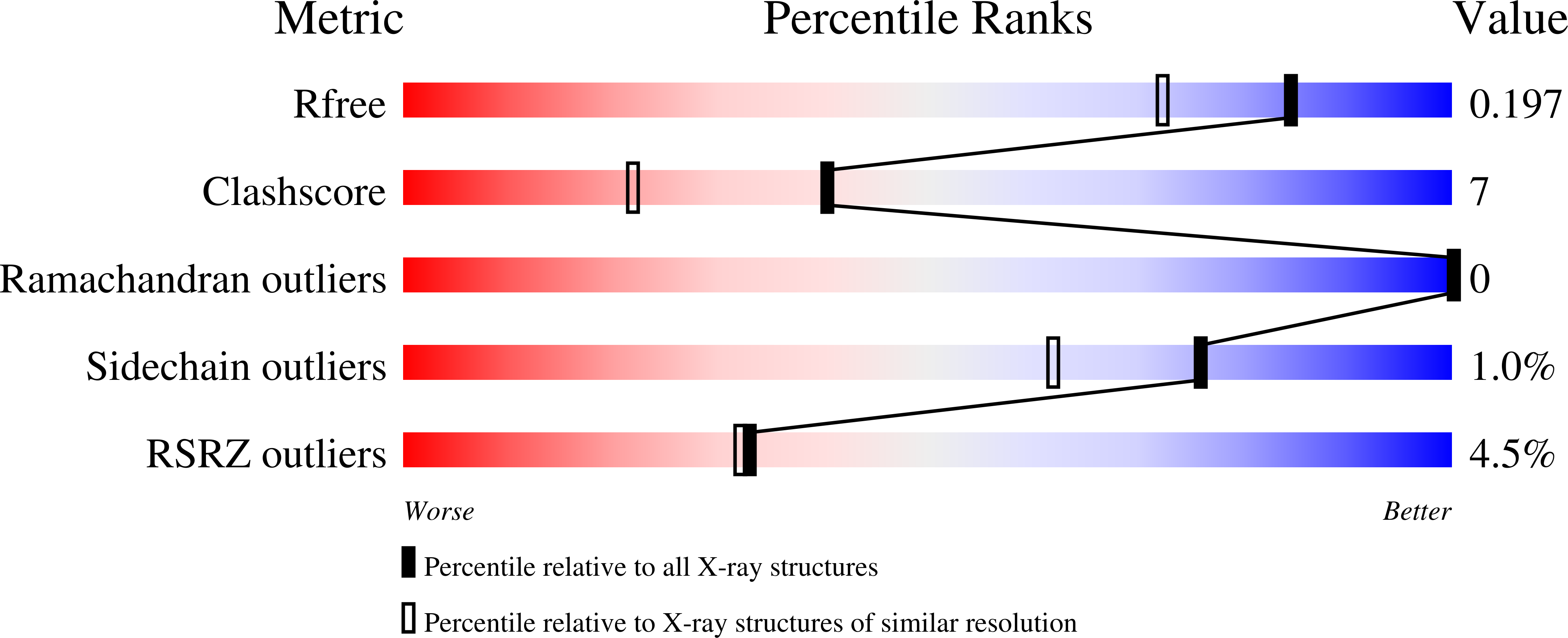
4ADN, 4ADO - PubMed Abstract:
Fusidic acid (FA) is a bacteriostatic antibiotic that locks elongation factor G (EF-G) to the ribosome after GTP hydrolysis during elongation and ribosome recycling. The plasmid pUB101-encoded protein FusB causes FA resistance in clinical isolates of Staphylococcus aureus through an interaction with EF-G. Here, we report 1.6 and 2.3 Å crystal structures of FusB. We show that FusB is a two-domain protein lacking homology to known structures, where the N-terminal domain is a four-helix bundle and the C-terminal domain has an alpha/beta fold containing a C4 treble clef zinc finger motif and two loop regions with conserved basic residues. Using hybrid constructs between S. aureus EF-G that binds to FusB and Escherichia coli EF-G that does not, we show that the sequence determinants for FusB recognition reside in domain IV and involve the C-terminal helix of S. aureus EF-G. Further, using kinetic assays in a reconstituted translation system, we demonstrate that FusB can rescue FA inhibition of tRNA translocation as well as ribosome recycling. We propose that FusB rescues S. aureus from FA inhibition by preventing formation or facilitating dissociation of the FA-locked EF-G-ribosome complex.
Organizational Affiliation:
Department of Cell and Molecular Biology, BMC, P.O. Box 596, SE 751 24, Uppsala, Sweden.