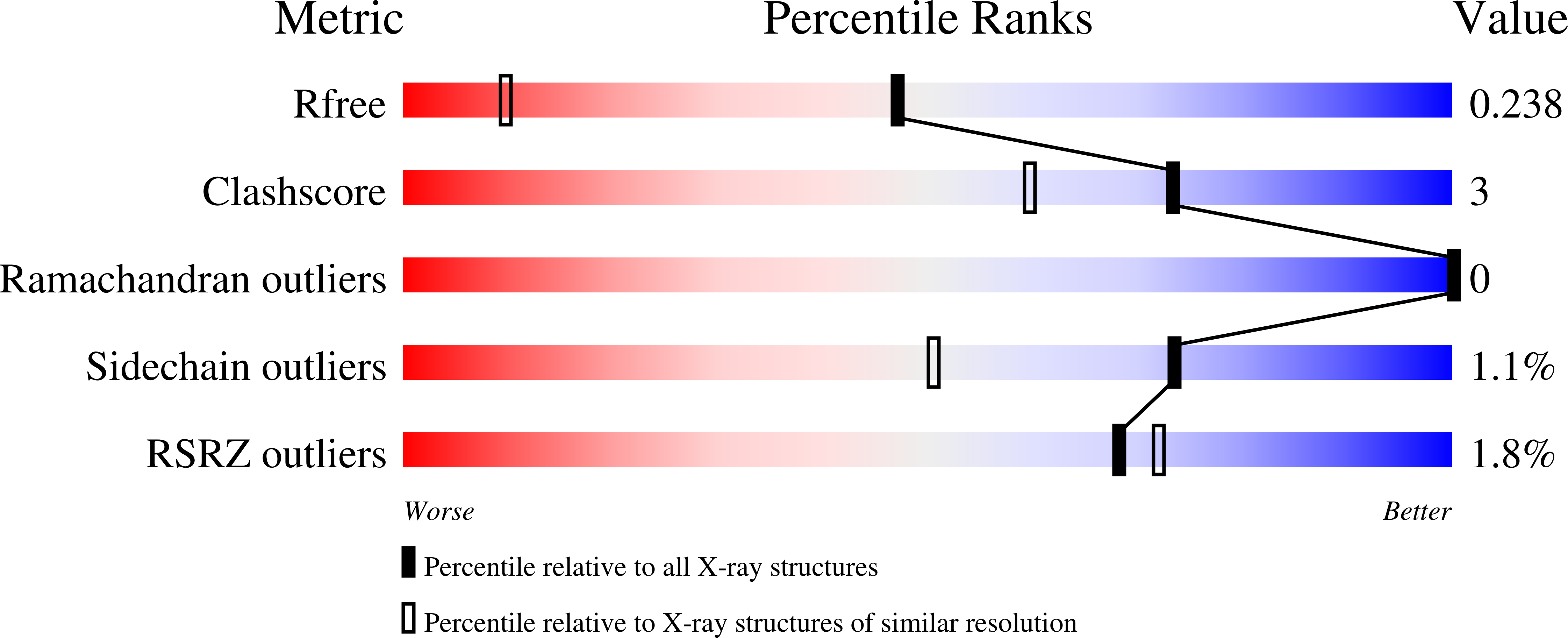
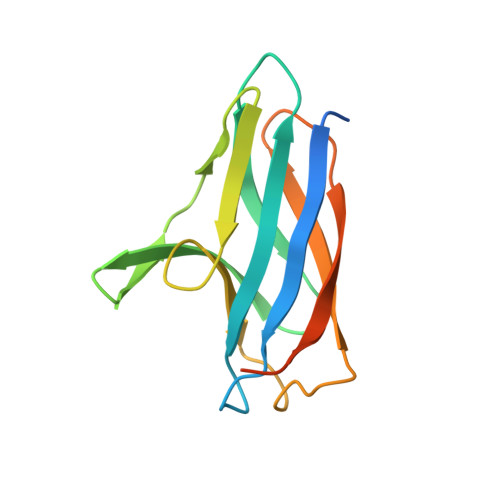
Structural and Mechanistic Insights Into an Extracytoplasmic Copper Trafficking Pathway in Streptomyces Lividans.
Blundell, K.L.I.M., Hough, M.A., Vijgenboom, E., Worrall, J.A.R.(2014) Biochem J 459: 525
- PubMed: 24548299
- DOI: https://doi.org/10.1042/BJ20140017
- Primary Citation of Related Structures:
3ZJA, 3ZK0, 4BPY - PubMed Abstract:
In Streptomyces lividans an extracytoplasmic copper-binding Sco protein plays a role in two unlinked processes: (i) initiating a morphological development switch and (ii) facilitating the co-factoring of the CuA domain of CcO (cytochrome c oxidase). How Sco obtains copper once secreted to the extracytoplasmic environment is unknown. In the present paper we report on a protein possessing an HX₆MX₂₁HXM motif that binds a single cuprous ion with subfemtomolar affinity. High-resolution X-ray structures of this extracytoplasmic copper chaperone-like protein (ECuC) in the apo- and Cu(I)-bound states reveal that the latter possesses a surface-accessible cuprous-ion-binding site located in a dish-shaped region of β-sheet structure. A cuprous ion is transferred under a favourable thermodynamic gradient from ECuC to Sco with no back transfer occurring. The ionization properties of the cysteine residues in the Cys⁸⁶xxxCys⁹⁰ copper-binding motif of Sco, together with their positional locations identified from an X-ray structure of Sco, suggests a role for Cys⁸⁶ in initiating an inter-complex ligand-exchange reaction with Cu(I)-ECuC. Generation of the genetic knockouts, Δsco, Δecuc and Δsco/ecuc, and subsequent in vivo assays lend support to the existence of a branched extracytoplasmic copper-trafficking pathway in S. lividans. One branch requires both Sco and to a certain extent ECuC to cofactor the CuA domain, whereas the other uses only Sco to deliver copper to a cuproenzyme to initiate morphological development.
Organizational Affiliation:
*School of Biological Sciences, University of Essex, Wivenhoe Park, Colchester CO4 3SQ, U.K.