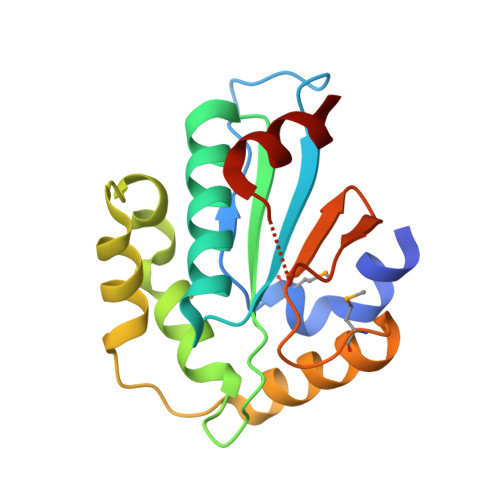
Structural insight into substrate recognition by the endoplasmic reticulum folding-sensor enzyme: crystal structure of third thioredoxin-like domain of UDP-glucose:glycoprotein glucosyltransferase
Zhu, T., Satoh, T., Kato, K.(2014) Sci Rep 4: 7322-7322
- PubMed: 25471383
- DOI: https://doi.org/10.1038/srep07322
- Primary Citation of Related Structures:
3WZS, 3WZT - PubMed Abstract:
The endoplasmic reticulum (ER) possesses a protein quality control system that supports the efficient folding of newly synthesized glycoproteins. In this system, a series of N-linked glycan intermediates displayed on proteins serve as quality tags. The ER folding-sensor enzyme UDP-glucose:glycoprotein glucosyltransferase (UGGT) operates as the gatekeeper for ER quality control by specifically transferring monoglucose residues to incompletely folded glycoproteins, thereby allowing them to interact with lectin chaperone complexes to facilitate their folding. Despite its functional importance, no structural information is available for this key enzyme to date. To elucidate the folding-sensor mechanism in the ER, we performed a structural study of UGGT. Based on bioinformatics analyses, the folding-sensor region of UGGT was predicted to harbour three tandem thioredoxin (Trx)-like domains, which are often found in proteins involved in ER quality control. Furthermore, we determined the three-dimensional structure of the third Trx-like domain, which exhibits an extensive hydrophobic patch concealed by its flexible C-terminal helix. Our structural data suggest that this hydrophobic patch is involved in intermolecular interactions, thereby contributing to the folding-sensor mechanism of UGGT.
Organizational Affiliation:
1] School of Physical Sciences, The Graduate University for Advanced Studies, 5-1 Higashiyama, Myodaiji, Okazaki, Aichi 444-8787, Japan [2] Okazaki Institute for Integrative Bioscience and Institute for Molecular Science, National Institutes of Natural Sciences, 5-1 Higashiyama, Myodaiji, Okazaki, Aichi 444-8787, Japan [3] Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan.