Rational design of heterodimeric protein using domain swapping for myoglobin.
Lin, Y.W., Nagao, S., Zhang, M., Shomura, Y., Higuchi, Y., Hirota, S.(2015) Angew Chem Int Ed Engl 54: 511-515
- PubMed: 25370865
- DOI: https://doi.org/10.1002/anie.201409267
- Primary Citation of Related Structures:
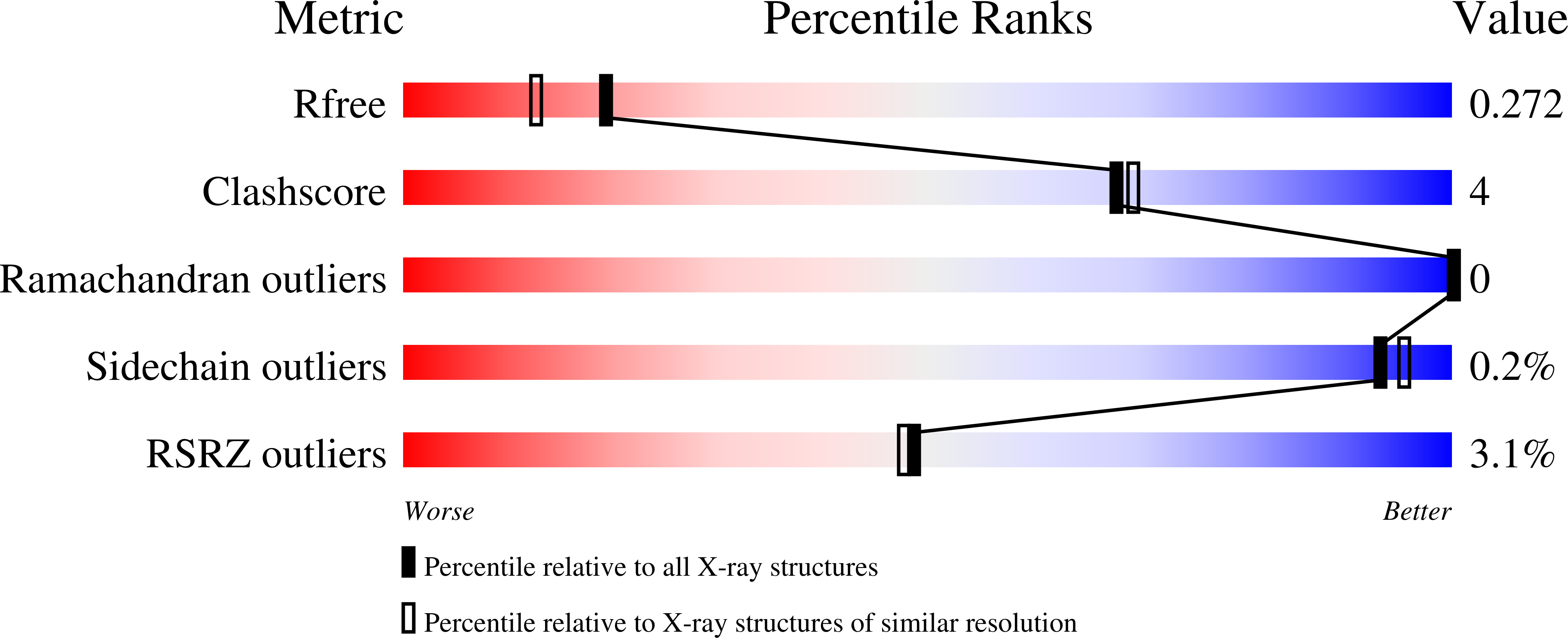
3WYO - PubMed Abstract:
Protein design is a useful method to create novel artificial proteins. A rational approach to design a heterodimeric protein using domain swapping for horse myoglobin (Mb) was developed. As confirmed by X-ray crystallographic analysis, a heterodimeric Mb with two different active sites was produced efficiently from two surface mutants of Mb, in which the charges of two amino acids involved in the dimer salt bridges were reversed in each mutant individually, with the active site of one mutant modified. This study shows that the method of constructing heterodimeric Mb with domain swapping is useful for designing artificial multiheme proteins.
Organizational Affiliation:
Graduate School of Materials Science, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara, 630-0192 (Japan); School of Chemistry and Chemical Engineering, University of South China, Hengyang 421001 (China).