Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
Akiyoshi, Y., Nakakura, Y., Hamada, D., Goda, N., Tenno, T., Narita, H., Nakagawa, A., Furuse, M., Suzuki, M., Hiroaki, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| E3 ubiquitin-protein ligase LNX | 94 | Mus musculus | Mutation(s): 0 Gene Names: Lnx1 | 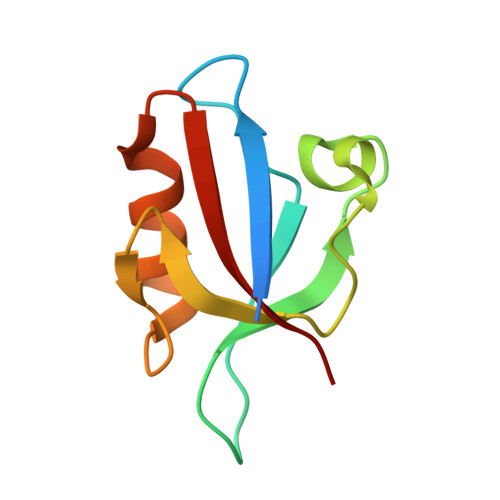 | |
UniProt | |||||
Find proteins for O70263 (Mus musculus) Explore O70263 Go to UniProtKB: O70263 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O70263 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.493 | α = 90 |
| b = 51.21 | β = 120 |
| c = 33.291 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |