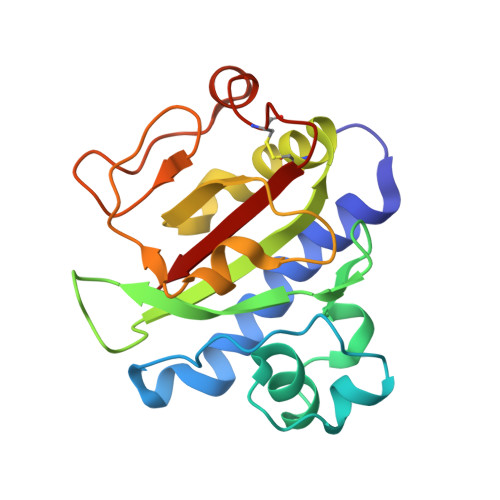
Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Fukakusa, S., Kawahara, K., Nakamura, S., Iwashita, T., Baba, S., Nishimura, M., Kobayashi, Y., Honda, T., Iida, T., Taniguchi, T., Ohkubo, T.(2012) Acta Crystallogr D Biol Crystallogr 68: 1418-1429
- PubMed: 22993096
- DOI: https://doi.org/10.1107/S0907444912034464
- Primary Citation of Related Structures:
3VOR - PubMed Abstract:
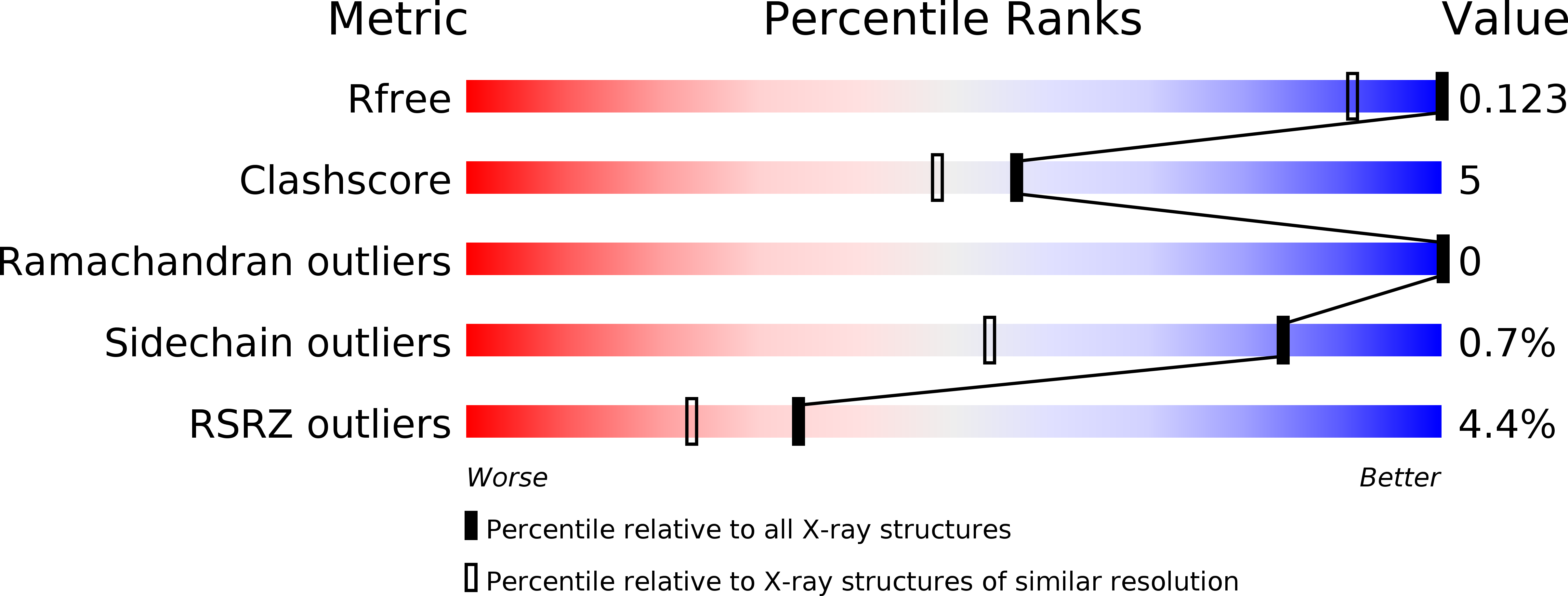
CofA, a major pilin subunit of colonization factor antigen III (CFA/III), forms pili that mediate small-intestinal colonization by enterotoxigenic Escherichia coli (ETEC). In this study, the crystal structure of an N-terminally truncated version of CofA was determined by single-wavelength anomalous diffraction (SAD) phasing using five sulfurs in the protein. Given the counterbalance between anomalous signal strength and the undesired X-ray absorption of the solvent, diffraction data were collected at 1.5 Å resolution using synchrotron radiation. These data were sufficient to elucidate the sulfur substructure at 1.38 Å resolution. The low solvent content (29%) of the crystal necessitated that density modification be performed with an additional 0.9 Å resolution data set to reduce the phase error caused by the small sulfur anomalous signal. The CofA structure showed the αβ-fold typical of type IVb pilins and showed high structural homology to that of TcpA for toxin-coregulated pili of Vibrio cholerae, including spatial distribution of key residues critical for pilin self-assembly. A pilus-filament model of CofA was built by computational docking and molecular-dynamics simulation using the previously reported filament model of TcpA as a structural template. This model revealed that the CofA filament surface was highly negatively charged and that a 23-residue-long loop between the α1 and α2 helices filled the gap between the pilin subunits. These characteristics could provide a unique binding epitope for the CFA/III pili of ETEC compared with other type IVb pili.
Organizational Affiliation:
Department of Pharmaceutical Sciences, Osaka University, 1-6 Yamadaoka, Suita, Osaka 565-0871, Japan.