Chiral-substrate-assisted stereoselective epoxidation catalyzed by H2O2-dependent cytochrome P450SP alpha
Fujishiro, T., Shoji, O., Kawakami, N., Watanabe, T., Sugimoto, H., Shiro, Y., Watanabe, Y.(2012) Chem Asian J 7: 2286-2293
- PubMed: 22700535
- DOI: https://doi.org/10.1002/asia.201200250
- Primary Citation of Related Structures:
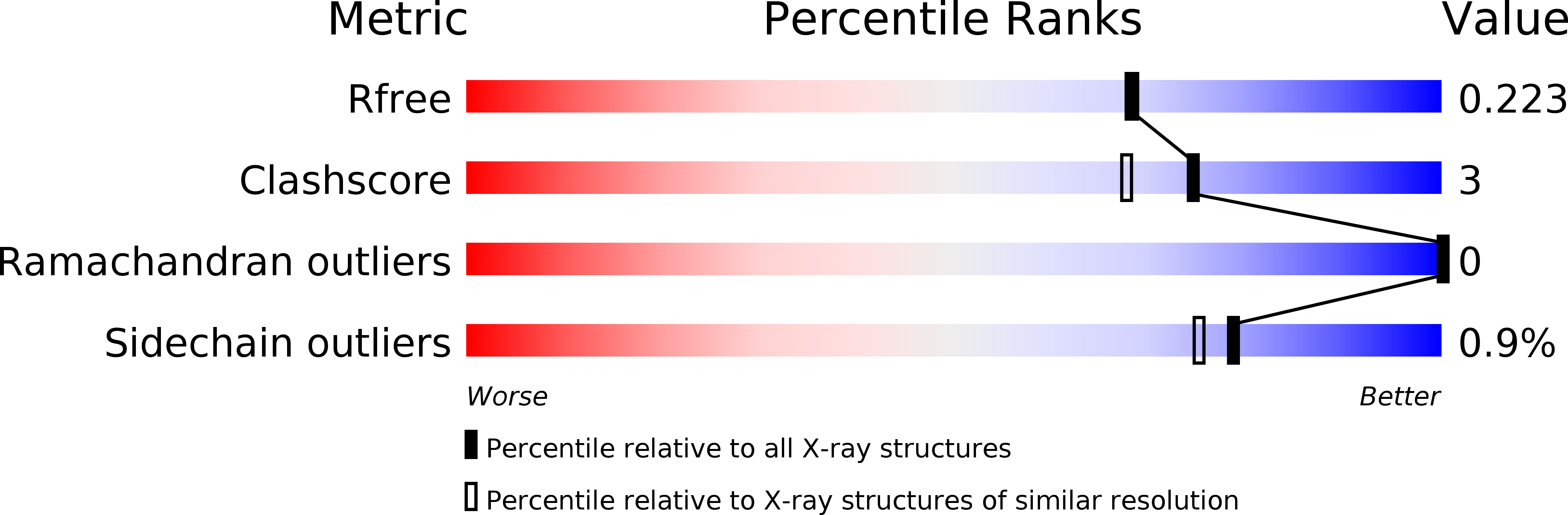
3VM4 - PubMed Abstract:
The stereoselective epoxidation of styrene was catalyzed by H(2) O(2) -dependent cytochrome P450(SPα) in the presence of carboxylic acids as decoy molecules. The stereoselectivity of styrene oxide could be altered by the nature of the decoy molecules. In particular, the chirality at the α-positions of the decoy molecules induced a clear difference in the chirality of the product: (R)-ibuprofen enhanced the formation of (S)-styrene oxide, whereas (S)-ibuprofen preferentially afforded (R)-styrene oxide. The crystal structure of an (R)-ibuprofen-bound cytochrome P450(SPα) (resolution 1.9 Å) revealed that the carboxylate group of (R)-ibuprofen served as an acid-base catalyst to initiate the epoxidation. A docking simulation of the binding of styrene in the active site of the (R)-ibuprofen-bound form suggested that the orientation of the vinyl group of styrene in the active site agreed with the formation of (S)-styrene oxide.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8602, Japan.