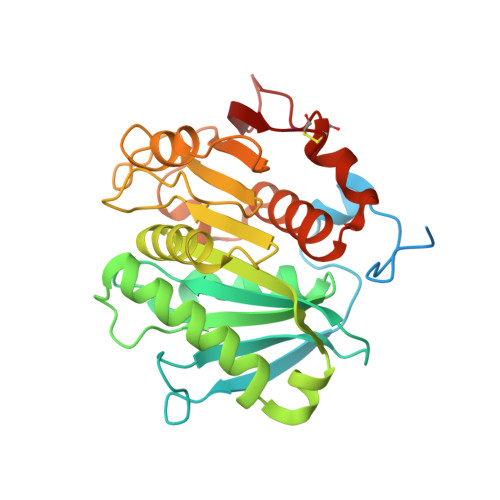
Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
Kitadokoro, K., Thumarat, U., Nakamura, R., Nishimura, K., Karatani, H., Suzuki, H., Kawai, F.(2012) Polym Degrad Stab 97: 771-775