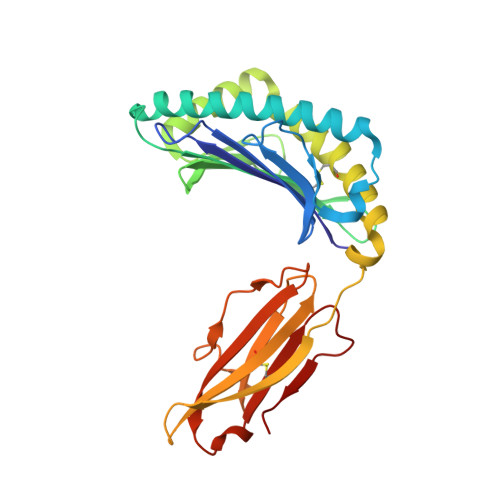
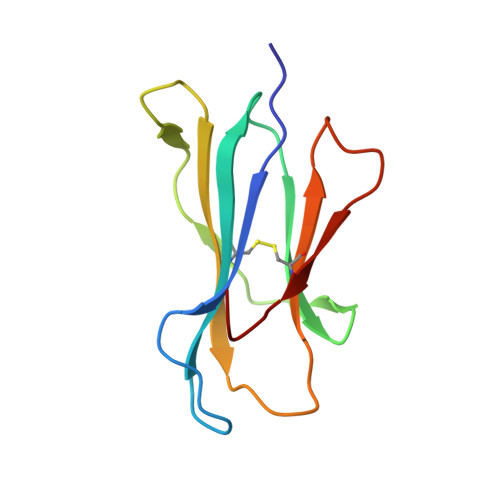
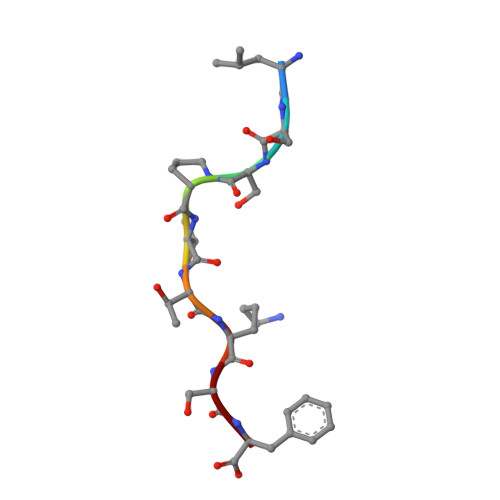
Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B
Vivian, J.P., Duncan, R.C., Berry, R., O'Connor, G.M., Reid, H.H., Beddoe, T., Gras, S., Saunders, P.M., Olshina, M.A., Widjaja, J.M.L., Harpur, C.M., Lin, J., Maloveste, S.M., Price, D.A., Lafont, B.A.P., McVicar, D.W., Clements, C.S., Brooks, A.G., Rossjohn, J.(2011) Nature 479: 401-405
- PubMed: 22020283
- DOI: https://doi.org/10.1038/nature10517
- Primary Citation of Related Structures:
3VH8 - PubMed Abstract:
Members of the killer cell immunoglobulin-like receptor (KIR) family, a large group of polymorphic receptors expressed on natural killer (NK) cells, recognize particular peptide-laden human leukocyte antigen (pHLA) class I molecules and have a pivotal role in innate immune responses. Allelic variation and extensive polymorphism within the three-domain KIR family (KIR3D, domains D0-D1-D2) affects pHLA binding specificity and is linked to the control of viral replication and the treatment outcome of certain haematological malignancies. Here we describe the structure of a human KIR3DL1 receptor bound to HLA-B*5701 complexed with a self-peptide. KIR3DL1 clamped around the carboxy-terminal end of the HLA-B*5701 antigen-binding cleft, resulting in two discontinuous footprints on the pHLA. First, the D0 domain, a distinguishing feature of the KIR3D family, extended towards β2-microglobulin and abutted a region of the HLA molecule with limited polymorphism, thereby acting as an 'innate HLA sensor' domain. Second, whereas the D2-HLA-B*5701 interface exhibited a high degree of complementarity, the D1-pHLA-B*5701 contacts were suboptimal and accommodated a degree of sequence variation both within the peptide and the polymorphic region of the HLA molecule. Although the two-domain KIR (KIR2D) and KIR3DL1 docked similarly onto HLA-C and HLA-B respectively, the corresponding D1-mediated interactions differed markedly, thereby providing insight into the specificity of KIR3DL1 for discrete HLA-A and HLA-B allotypes. Collectively, in association with extensive mutagenesis studies at the KIR3DL1-pHLA-B*5701 interface, we provide a framework for understanding the intricate interplay between peptide variability, KIR3D and HLA polymorphism in determining the specificity requirements of this essential innate interaction that is conserved across primate species.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.