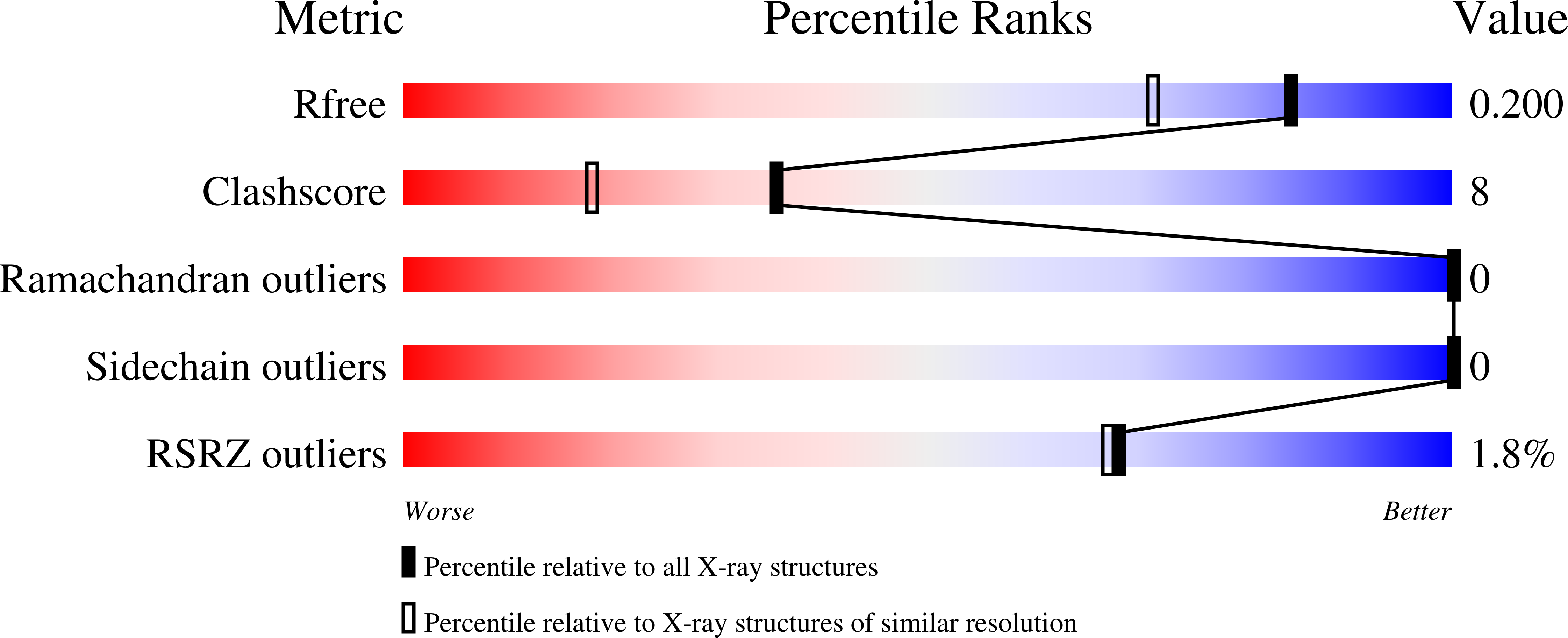
Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
Barabas, O., Nemeth, V., Bodor, A., Perczel, A., Rosta, E., Kele, Z., Zagyva, I., Szabadka, Z., Grolmusz, V.I., Wilmanns, M., Vertessy, B.G.To be published.