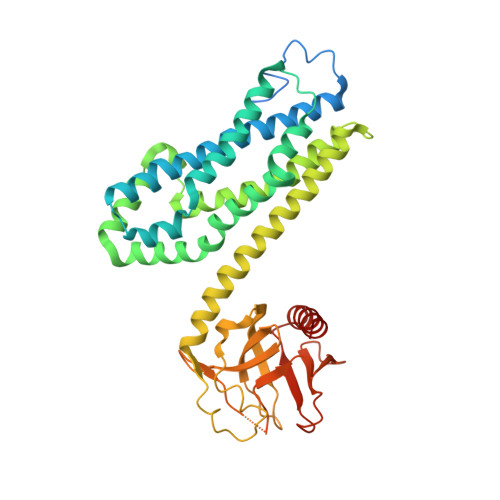
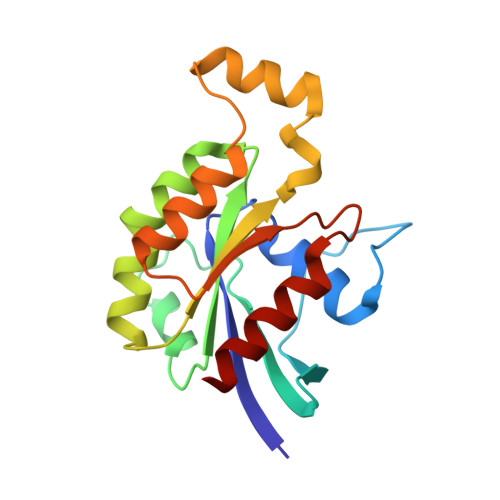
Insights into the Molecular Activation Mechanism of the RhoA-specific Guanine Nucleotide Exchange Factor, PDZRhoGEF.
Bielnicki, J.A., Shkumatov, A.V., Derewenda, U., Somlyo, A.V., Svergun, D.I., Derewenda, Z.S.(2011) J Biol Chem 286: 35163-35175
- PubMed: 21816819
- DOI: https://doi.org/10.1074/jbc.M111.270918
- Primary Citation of Related Structures:
3T06 - PubMed Abstract:
PDZRhoGEF (PRG) belongs to a small family of RhoA-specific nucleotide exchange factors that mediates signaling through select G-protein-coupled receptors via Gα(12/13) and activates RhoA by catalyzing the exchange of GDP to GTP. PRG is a multidomain protein composed of PDZ, regulators of G-protein signaling-like (RGSL), Dbl-homology (DH), and pleckstrin-homology (PH) domains. It is autoinhibited in cytosol and is believed to undergo a conformational rearrangement and translocation to the membrane for full activation, although the molecular details of the regulation mechanism are not clear. It has been shown recently that the main autoregulatory elements of PDZRhoGEF, the autoinhibitory "activation box" and the "GEF switch," which is required for full activation, are located directly upstream of the catalytic DH domain and its RhoA binding surface, emphasizing the functional role of the RGSL-DH linker. Here, using a combination of biophysical and biochemical methods, we show that the mechanism of PRG regulation is yet more complex and may involve an additional autoinhibitory element in the form of a molten globule region within the linker between RGSL and DH domains. We propose a novel, two-tier model of autoinhibition where the activation box and the molten globule region act synergistically to impair the ability of RhoA to bind to the catalytic DH-PH tandem. The molten globule region and the activation box become less ordered in the PRG-RhoA complex and dissociate from the RhoA-binding site, which may constitute a critical step leading to PRG activation.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics University of Virginia, Charlottesville, Virginia 22908, USA.