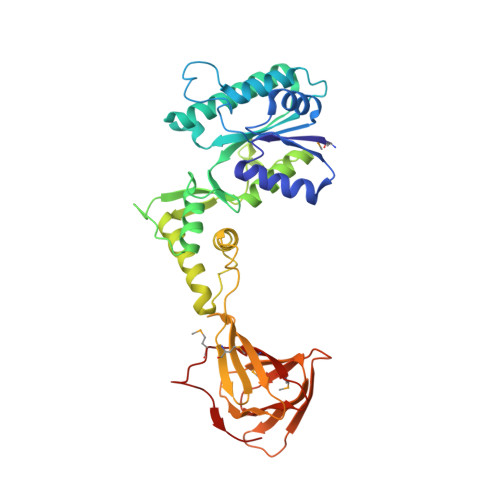
Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Miyafusa, T., Caaveiro, J.M., Tanaka, Y., Tsumoto, K.(2012) Biochem J 443: 671-680
- PubMed: 22320426
- DOI: https://doi.org/10.1042/BJ20112049
- Primary Citation of Related Structures:
3ST7, 3VHR - PubMed Abstract:
CP (capsular polysaccharide) is an important virulence factor during infections by the bacterium Staphylococcus aureus. The enzyme CapF is an attractive therapeutic candidate belonging to the biosynthetic route of CP of pathogenic strains of S. aureus. In the present study, we report two independent crystal structures of CapF in an open form of the apoenzyme. CapF is a homodimer displaying a characteristic dumb-bell-shaped architecture composed of two domains. The N-terminal domain (residues 1-252) adopts a Rossmann fold belonging to the short-chain dehydrogenase/reductase family of proteins. The C-terminal domain (residues 252-369) displays a standard cupin fold with a Zn2+ ion bound deep in the binding pocket of the β-barrel. Functional and thermodynamic analyses indicated that each domain catalyses separate enzymatic reactions. The cupin domain is necessary for the C3-epimerization of UDP-4-hexulose. Meanwhile, the N-terminal domain catalyses the NADPH-dependent reduction of the intermediate species generated by the cupin domain. Analysis by ITC (isothermal titration calorimetry) revealed a fascinating thermodynamic switch governing the attachment and release of the coenzyme NADPH during each catalytic cycle. These observations suggested that the binding of coenzyme to CapF facilitates a disorder-to-order transition in the catalytic loop of the reductase (N-terminal) domain. We anticipate that the present study will improve the general understanding of the synthesis of CP in S. aureus and will aid in the design of new therapeutic agents against this pathogenic bacterium.
Organizational Affiliation:
Department of Medical Genome Science, School of Frontier Sciences, The University of Tokyo, Minato-ku, Tokyo 108-8639, Japan.