Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN)
Yuzawa, S., Kamakura, S., Iwakiri, Y., Hayase, J., Sumimoto, H.(2011) Proc Natl Acad Sci U S A 108: 19210-19215
- PubMed: 22074847
- DOI: https://doi.org/10.1073/pnas.1110951108
- Primary Citation of Related Structures:
3SF4 - PubMed Abstract:
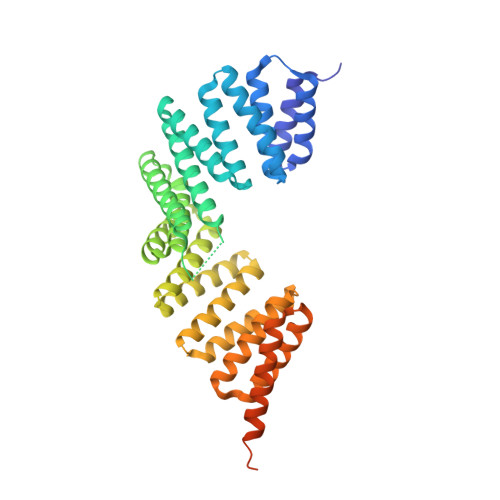
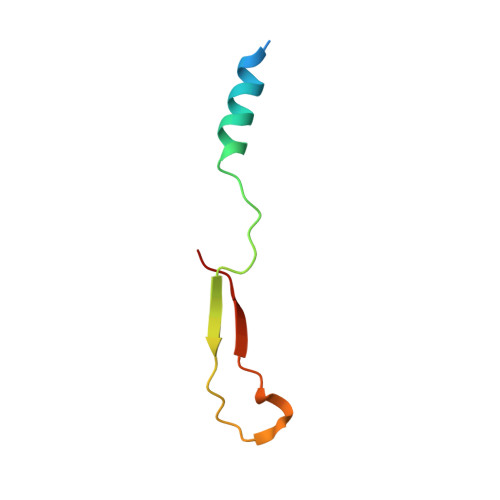
Interaction between the mammalian cell polarity proteins mInsc (mammalian homologue of Inscuteable) and Leu-Gly-Asn repeat-enriched protein (LGN), as well as that between their respective Drosophila homologues Inscuteable and Partner of Inscuteable (Pins), plays crucial roles in mitotic spindle orientation, a process contributing to asymmetric cell division. Here, we report a crystal structure of the LGN-binding domain (LBD) of human mInsc complexed with the N-terminal tetratricopeptide repeat (TPR) motifs of human LGN at 2.6-Å resolution. In the complex, mInsc-LBD adopts an elongated structure with three binding modules--an α-helix, an extended region, and a β-sheet connected with a loop--that runs antiparallel to LGN along the concave surface of the superhelix formed by the TPRs. Structural analysis and structure-based mutagenesis define residues that are critical for mInsc-LGN association, and reveal that the activator of G-protein signaling 3 (AGS3)-binding protein Frmpd1 [4.1/ezrin/radixin/moesin (FERM) and PSD-95/Dlg/ZO-1 (PDZ) domain-containing protein 1] and its relative Frmpd4 interact with LGN via a region homologous to a part of mInsc-LBD, whereas nuclear mitotic apparatus protein (NuMA) and the C terminus of LGN recognize the TPR domain in a manner different from that by mInsc. mInsc binds to LGN with the highest affinity (K(D) ≈ 2.4 nM) and effectively replaces the Frmpd proteins, NuMA, and the LGN C terminus, suggesting the priority of mInsc in binding to LGN. We also demonstrate, using mutant proteins, that mInsc-LGN interaction is vital for stabilization of LGN and for intracellular localization of mInsc.
Organizational Affiliation:
Department of Biochemistry, Kyushu University Graduate School of Medical Sciences, Fukuoka 812-8582, Japan.