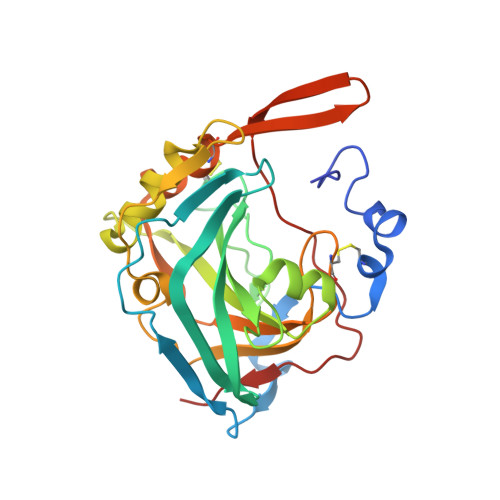
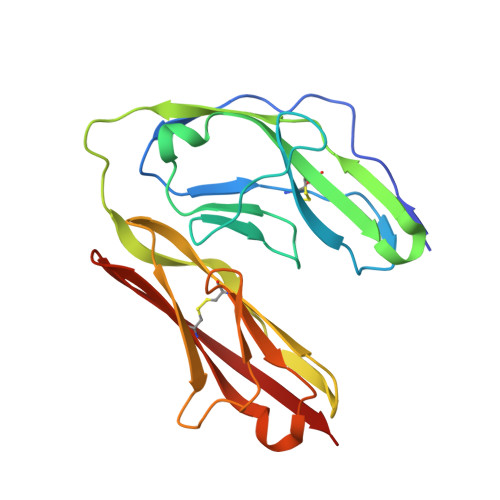
A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells.
Lamprianou, S., Chatzopoulou, E., Thomas, J.L., Bouyain, S., Harroch, S.(2011) Proc Natl Acad Sci U S A 108: 17498-17503
- PubMed: 21969550
- DOI: https://doi.org/10.1073/pnas.1108774108
- Primary Citation of Related Structures:
3S97 - PubMed Abstract:
The six members of the contactin (CNTN) family of neural cell adhesion molecules are involved in the formation and maintenance of the central nervous system (CNS) and have been linked to mental retardation and neuropsychiatric disorders such as autism. Five of the six CNTNs bind to the homologous receptor protein tyrosine phosphatases gamma (PTPRG) and zeta (PTPRZ), but the biological roles of these interactions remain unclear. We report here the cocrystal structure of the carbonic anhydrase-like domain of PTPRZ bound to tandem Ig repeats of CNTN1 and combine these structural data with binding assays to show that PTPRZ binds specifically to CNTN1 expressed at the surface of oligodendrocyte precursor cells. Furthermore, analyses of glial cell populations in wild-type and PTPRZ-deficient mice show that the binding of PTPRZ to CNTN1 expressed at the surface of oligodendrocyte precursor cells inhibits their proliferation and promotes their development into mature oligodendrocytes. Overall, these results implicate the PTPRZ/CNTN1 complex as a previously unknown modulator of oligodendrogenesis.
Organizational Affiliation:
Départment de Neuroscience, Institut Pasteur de Paris, 75624 Paris, France.