Structural and functional characterization of a noncanonical nucleoside triphosphate pyrophosphatase from Thermotoga maritima.
Awwad, K., Desai, A., Smith, C., Sommerhalter, M.(2013) Acta Crystallogr D Biol Crystallogr 69: 184-193
- PubMed: 23385455
- DOI: https://doi.org/10.1107/S0907444912044630
- Primary Citation of Related Structures:
3S86 - PubMed Abstract:
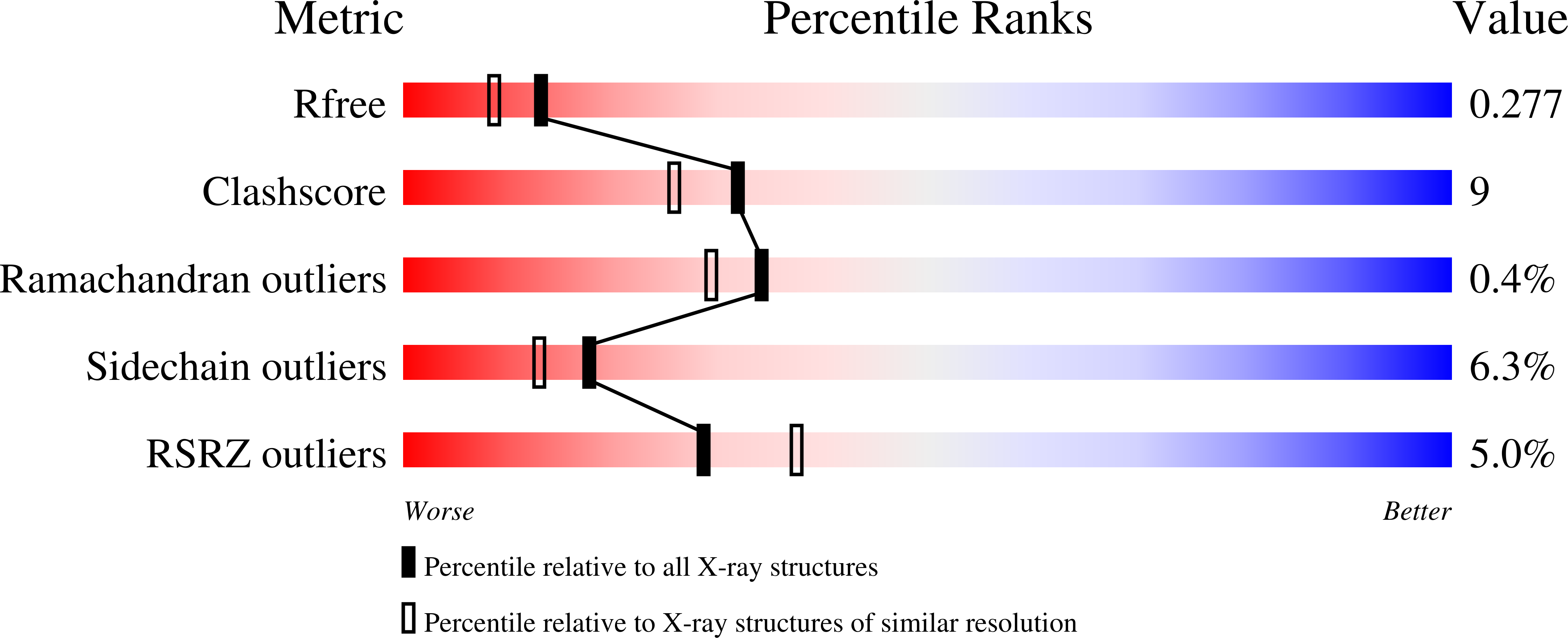
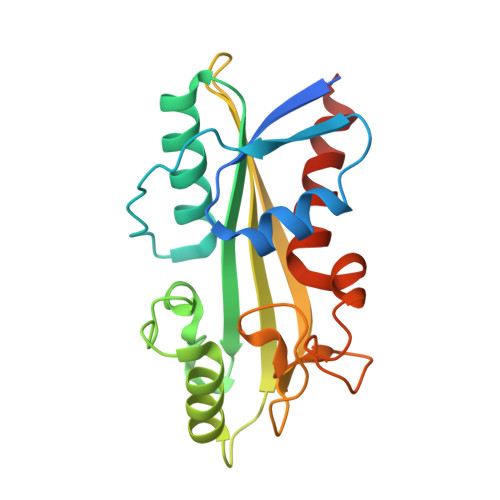
The hyperthermophilic bacterium Thermotoga maritima has a noncanonical nucleoside triphosphatase that catalyzes the conversion of inosine triphosphate (ITP), deoxyinosine triphosphate (dITP) and xanthosine triphosphate (XTP) into inosine monophosphate (IMP), deoxyinosine monophosphate (IMP) and xanthosine monophosphate (XMP), respectively. The k(cat)/K(m) values determined at 323 and 353 K fall between 1.31 × 10(4) and 7.80 × 10(4) M(-1) s(-1). ITP and dITP are slightly preferred over XTP. Activity towards canonical nucleoside triphosphates (ATP and GTP) was not detected. The enzyme has an absolute requirement for Mg(2+) as a cofactor and has a preference for alkaline conditions. A protein X-ray structure of the enzyme with bound IMP was obtained at 2.15 Å resolution. The active site houses a well conserved network of residues that are critical for substrate recognition and catalysis. The crystal structure shows a tetramer with two possible dimer interfaces. One of these interfaces strongly resembles the dimer interface that is found in the structures of other noncanonical nucleoside pyrophosphatases from human (human ITPase) and archaea (Mj0226 and PhNTPase).
Organizational Affiliation:
Chemistry and Biochemistry, California State University East Bay, 25800 Carlos Bee Boulevard, Hayward, CA 94542, USA.