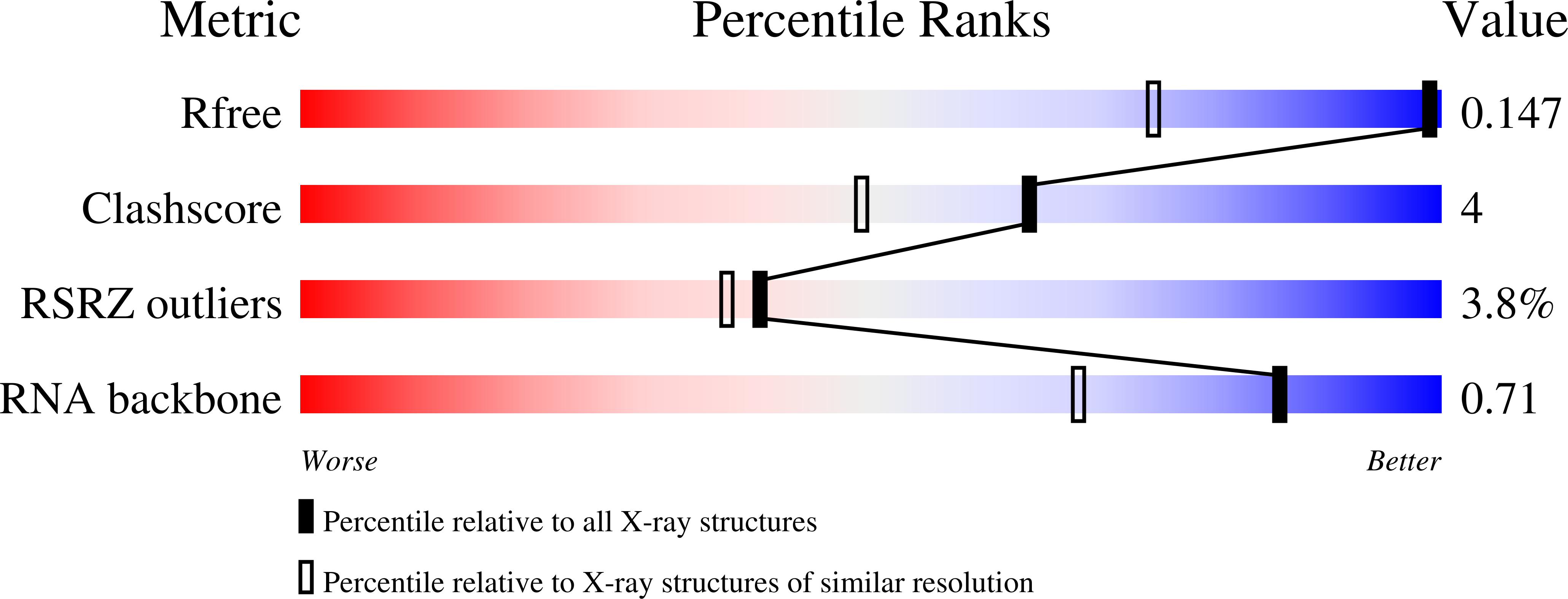2'-Azido RNA, a Versatile Tool for Chemical Biology: Synthesis, X-ray Structure, siRNA Applications, Click Labeling.
Fauster, K., Hartl, M., Santner, T., Aigner, M., Kreutz, C., Bister, K., Ennifar, E., Micura, R.(2012) ACS Chem Biol 7: 581-589
- PubMed: 22273279
- DOI: https://doi.org/10.1021/cb200510k
- Primary Citation of Related Structures:
3S7C, 3S8U - PubMed Abstract:
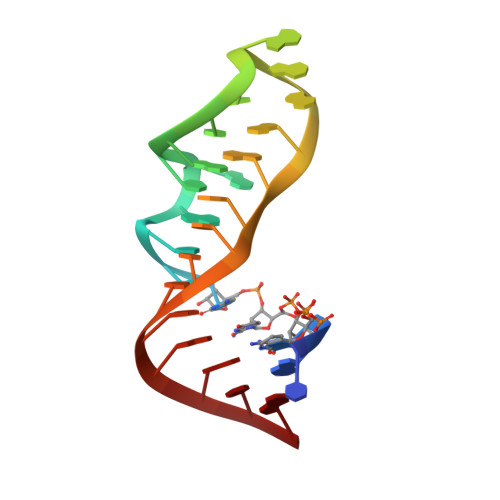
Chemical modification can significantly enrich the structural and functional repertoire of ribonucleic acids and endow them with new outstanding properties. Here, we report the syntheses of novel 2'-azido cytidine and 2'-azido guanosine building blocks and demonstrate their efficient site-specific incorporation into RNA by mastering the synthetic challenge of using phosphoramidite chemistry in the presence of azido groups. Our study includes the detailed characterization of 2'-azido nucleoside containing RNA using UV-melting profile analysis and CD and NMR spectroscopy. Importantly, the X-ray crystallographic analysis of 2'-azido uridine and 2'-azido adenosine modified RNAs reveals crucial structural details of this modification within an A-form double helical environment. The 2'-azido group supports the C3'-endo ribose conformation and shows distinct water-bridged hydrogen bonding patterns in the minor groove. Additionally, siRNA induced silencing of the brain acid soluble protein (BASP1) encoding gene in chicken fibroblasts demonstrated that 2'-azido modifications are well tolerated in the guide strand, even directly at the cleavage site. Furthermore, the 2'-azido modifications are compatible with 2'-fluoro and/or 2'-O-methyl modifications to achieve siRNAs of rich modification patterns and tunable properties, such as increased nuclease resistance or additional chemical reactivity. The latter was demonstrated by the utilization of the 2'-azido groups for bioorthogonal Click reactions that allows efficient fluorescent labeling of the RNA. In summary, the present comprehensive investigation on site-specifically modified 2'-azido RNA including all four nucleosides provides a basic rationale behind the physico- and biochemical properties of this flexible and thus far neglected type of RNA modification.
Organizational Affiliation:
Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, 6020 Innsbruck, Austria.