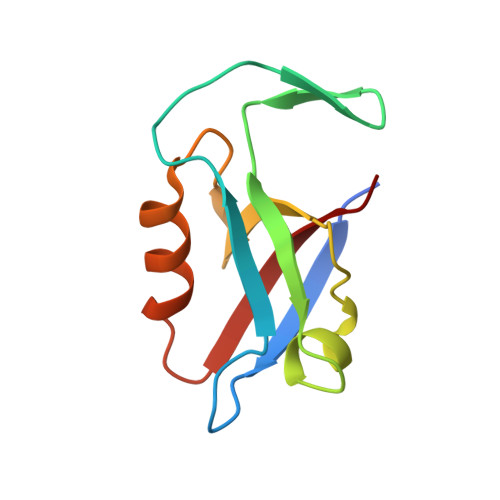
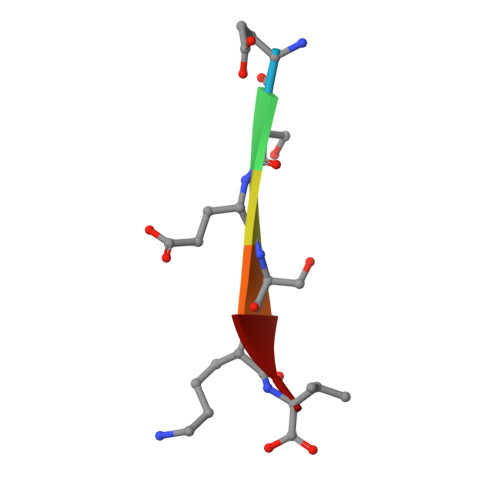
Mechanism underlying selective regulation of G protein-gated inwardly rectifying potassium channels by the psychostimulant-sensitive sorting nexin 27.
Balana, B., Maslennikov, I., Kwiatkowski, W., Stern, K.M., Bahima, L., Choe, S., Slesinger, P.A.(2011) Proc Natl Acad Sci U S A 108: 5831-5836
- PubMed: 21422294
- DOI: https://doi.org/10.1073/pnas.1018645108
- Primary Citation of Related Structures:
3QDO, 3QE1, 3QGL - PubMed Abstract:
G protein-gated inwardly rectifying potassium (GIRK) channels are important gatekeepers of neuronal excitability. The surface expression of neuronal GIRK channels is regulated by the psychostimulant-sensitive sorting nexin 27 (SNX27) protein through a class I (-X-Ser/Thr-X-Φ, where X is any residue and Φ is a hydrophobic amino acid) PDZ-binding interaction. The G protein-insensitive inward rectifier channel (IRK1) contains the same class I PDZ-binding motif but associates with a different synaptic PDZ protein, postsynaptic density protein 95 (PSD95). The mechanism by which SNX27 and PSD95 discriminate these channels was previously unclear. Using high-resolution structures coupled with biochemical and functional analyses, we identified key amino acids upstream of the channel's canonical PDZ-binding motif that associate electrostatically with a unique structural pocket in the SNX27-PDZ domain. Changing specific charged residues in the channel's carboxyl terminus or in the PDZ domain converts the selective association and functional regulation by SNX27. Elucidation of this unique interaction site between ion channels and PDZ-containing proteins could provide a therapeutic target for treating brain diseases.
Organizational Affiliation:
Clayton Foundation Laboratories for Peptide Biology, Salk Institute for Biological Studies, La Jolla, CA 92037, USA.