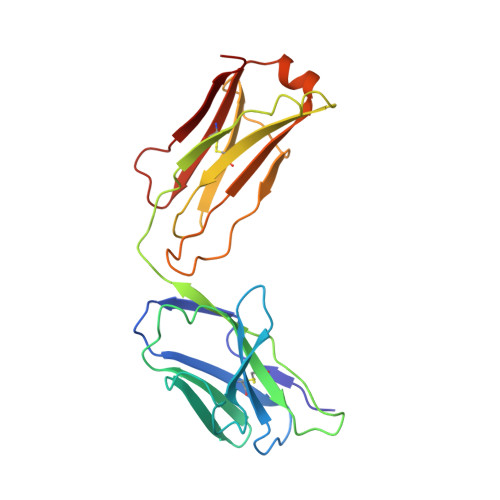
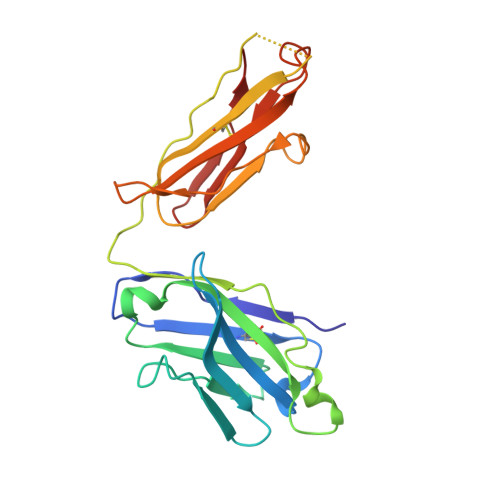
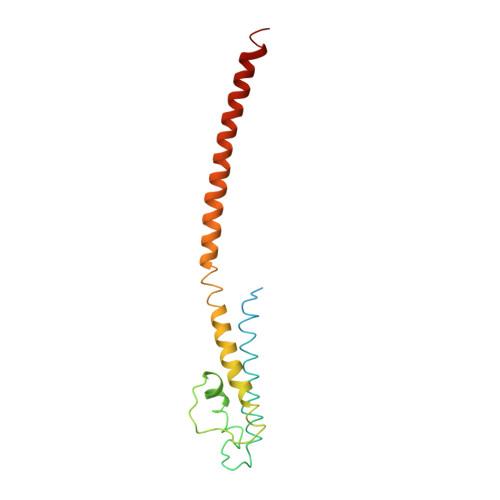
Mechanism of activation gating in the full-length KcsA K+ channel.
Uysal, S., Cuello, L.G., Cortes, D.M., Koide, S., Kossiakoff, A.A., Perozo, E.(2011) Proc Natl Acad Sci U S A 108: 11896-11899
- PubMed: 21730186
- DOI: https://doi.org/10.1073/pnas.1105112108
- Primary Citation of Related Structures:
3PJS - PubMed Abstract:
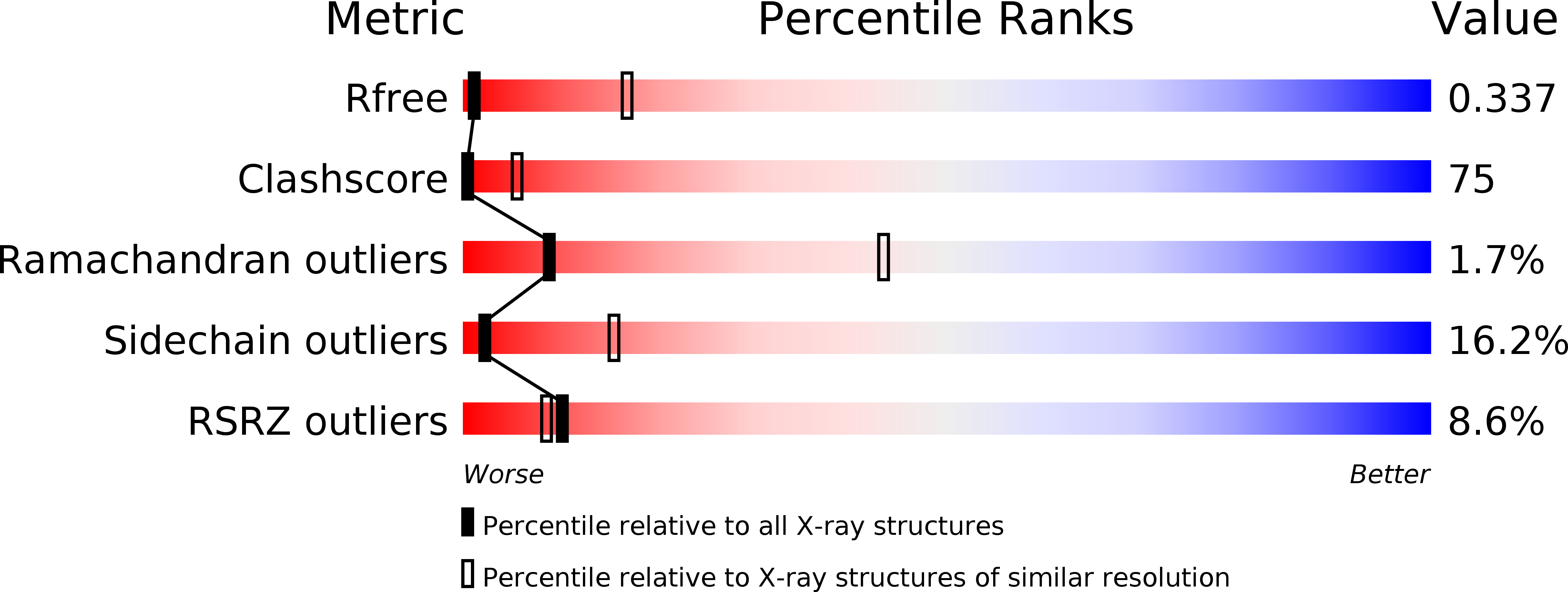
Using a constitutively active channel mutant, we solved the structure of full-length KcsA in the open conformation at 3.9 Å. The structure reveals that the activation gate expands about 20 Å, exerting a strain on the bulge helices in the C-terminal domain and generating side windows large enough to accommodate hydrated K(+) ions. Functional and spectroscopic analysis of the gating transition provides direct insight into the allosteric coupling between the activation gate and the selectivity filter. We show that the movement of the inner gate helix is transmitted to the C-terminus as a straightforward expansion, leading to an upward movement and the insertion of the top third of the bulge helix into the membrane. We suggest that by limiting the extent to which the inner gate can open, the cytoplasmic domain also modulates the level of inactivation occurring at the selectivity filter.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Institute for Biophysical Dynamics, University of Chicago, Chicago, IL 60637, USA.