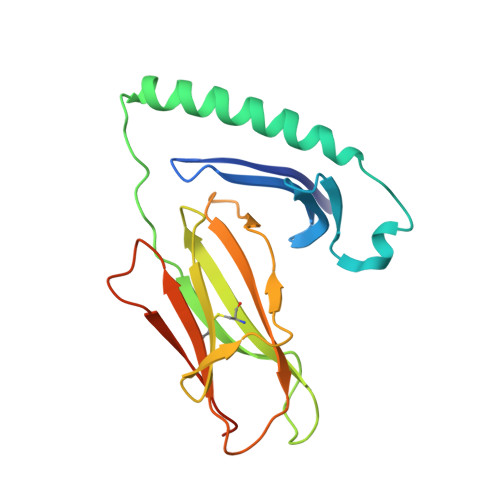
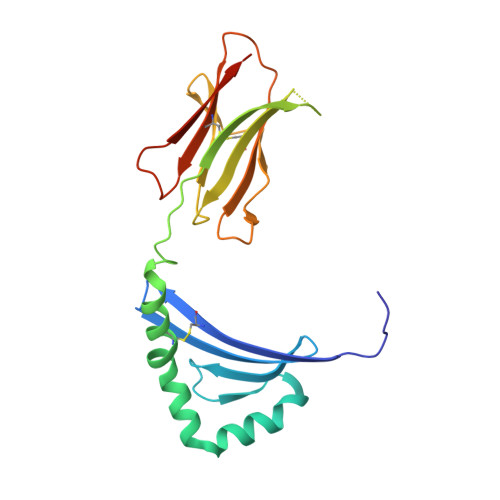
Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Gunther, S., Schlundt, A., Sticht, J., Roske, Y., Heinemann, U., Wiesmuller, K.H., Jung, G., Falk, K., Rotzschke, O., Freund, C.(2010) Proc Natl Acad Sci U S A 107: 22219-22224
- PubMed: 21115828
- DOI: https://doi.org/10.1073/pnas.1014708107
- Primary Citation of Related Structures:
3PDO, 3PGC, 3PGD - PubMed Abstract:
T-cell recognition of peptides bound to MHC class II (MHCII) molecules is a central event in cell-mediated adaptive immunity. The current paradigm holds that prebound class II-associated invariant chain peptides (CLIP) and all subsequent antigens maintain a canonical orientation in the MHCII binding groove. Here we provide evidence for MHCII-bound CLIP inversion. NMR spectroscopy demonstrates that the interconversion from the canonical to the inverse alignment is a dynamic process, and X-ray crystallography shows that conserved MHC residues form a hydrogen bond network with the peptide backbone in both orientations. The natural catalyst HLA-DM accelerates peptide reorientation and the exchange of either canonically or inversely bound CLIP against antigenic peptide. Thus, noncanonical MHC-CLIP displays the hallmarks of a structurally and functionally intact antigen-presenting complex.
Organizational Affiliation:
Leibniz-Institute for Molecular Pharmacology and Freie Universität Berlin, 13125 Berlin, Germany.