Discovery and structural characterization of a novel glycosidase family of marine origin.
Rebuffet, E., Groisillier, A., Thompson, A., Jeudy, A., Barbeyron, T., Czjzek, M., Michel, G.(2011) Environ Microbiol 13: 1253-1270
- PubMed: 21332624
- DOI: https://doi.org/10.1111/j.1462-2920.2011.02426.x
- Primary Citation of Related Structures:
3P2N - PubMed Abstract:
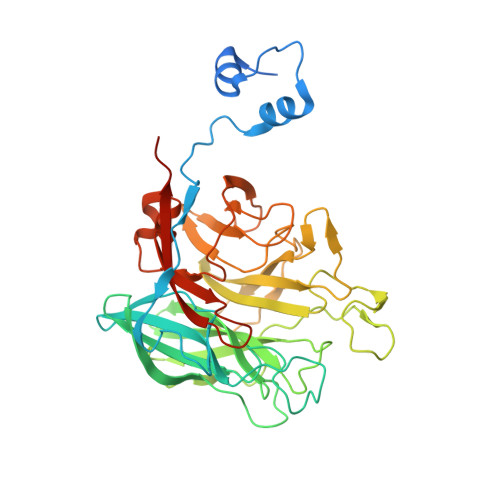
The genomic data on heterotrophic marine bacteria suggest the crucial role that microbes play in the global carbon cycle. However, the massive presence of hypothetical proteins hampers our understanding of the mechanisms by which this carbon cycle is carried out. Moreover, genomic data from marine microorganisms are essentially annotated in the light of the biochemical knowledge accumulated on bacteria and fungi which decompose terrestrial plants. However marine algal polysaccharides clearly differ from their terrestrial counterparts, and their associated enzymes usually constitute novel protein families. In this study, we have applied a combination of bioinformatics, targeted activity screening and structural biology to characterize a hypothetical protein from the marine bacterium Zobellia galactanivorans, which is distantly related to GH43 family. This protein is in fact a 1,3-α-3,6-anhydro-l-galactosidase (AhgA) which catalyses the last step in the degradation pathway of agars, a family of polysaccharides unique to red macroalgae. AhgA adopts a β-propeller fold and displays a zinc-dependent catalytic machinery. This enzyme is the first representative of a new family of glycoside hydrolases, especially abundant in coastal waters. Such genes of marine origin have been transferred to symbiotic microbes associated with marine fishes, but also with some specific human populations.
Organizational Affiliation:
UPMC University Paris 6 CNRS, UMR 7139 Marine Plants and Biomolecules, Station Biologique de Roscoff, Roscoff, Bretagne, France.