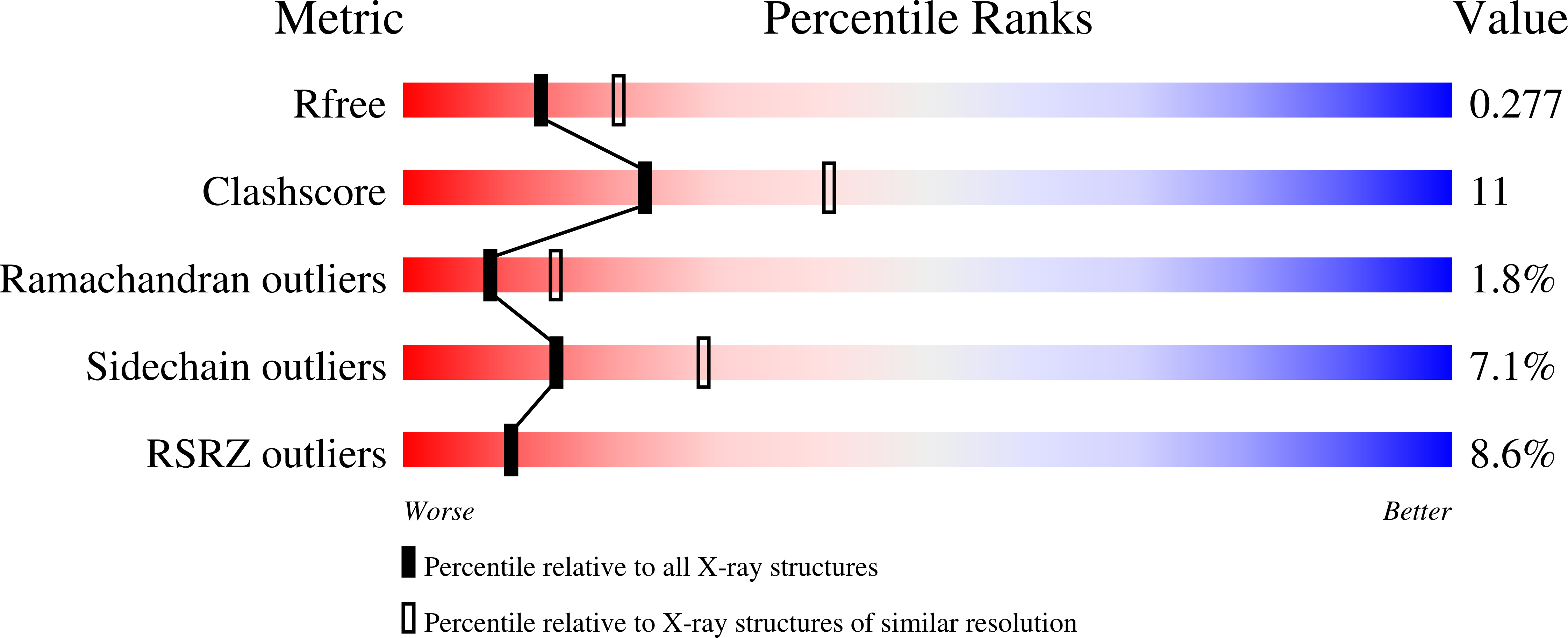
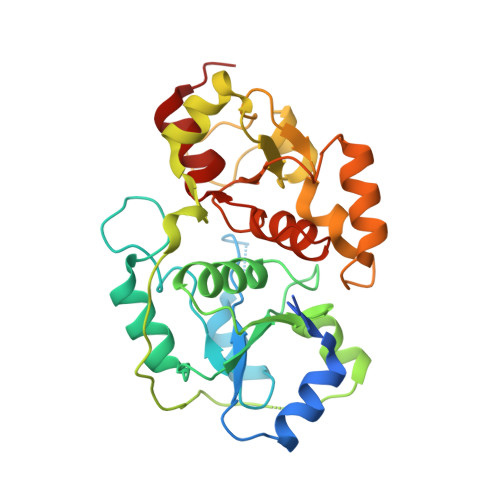
Human 3-Mercaptopyruvate Sulfurtransferase
Karlberg, T., Collins, R., Arrowsmith, C.H., Berglund, H., Bountra, C., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, I., Kotenyova, T., Kouznetsova, E., Moche, M., Nordlund, P., Nyman, T., Persson, C., Sehic, A., Schutz, P., Siponen, M.I., Thorsell, A.G., Tresaugues, L., Van Den Berg, S., Wahlberg, E., Weigelt, J., Welin, M., Schuler, H.To be published.