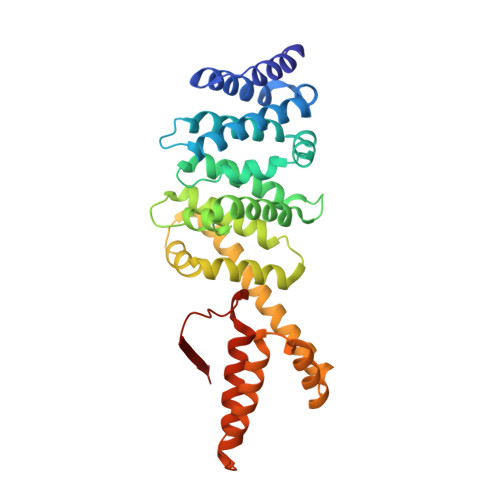
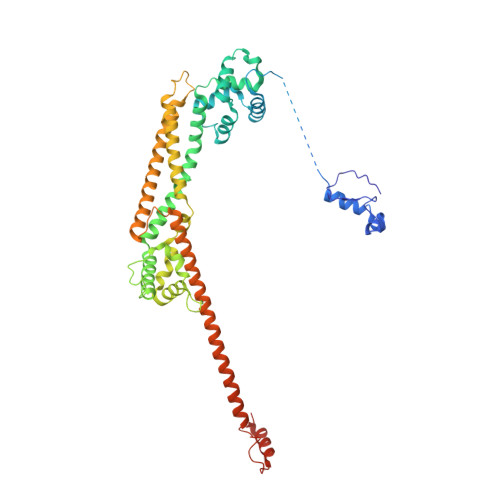
Crystal structure of the Formin mDia1 in autoinhibited conformation.
Otomo, T., Tomchick, D.R., Otomo, C., Machius, M., Rosen, M.K.(2010) PLoS One 5: e12896-e12896
- PubMed: 20927343
- DOI: https://doi.org/10.1371/journal.pone.0012896
- Primary Citation of Related Structures:
3OBV - PubMed Abstract:
Formin proteins utilize a conserved formin homology 2 (FH2) domain to nucleate new actin filaments. In mammalian diaphanous-related formins (DRFs) the FH2 domain is inhibited through an unknown mechanism by intramolecular binding of the diaphanous autoinhibitory domain (DAD) and the diaphanous inhibitory domain (DID). Here we report the crystal structure of a complex between DID and FH2-DAD fragments of the mammalian DRF, mDia1 (mammalian diaphanous 1 also called Drf1 or p140mDia). The structure shows a tetrameric configuration (4 FH2 + 4 DID) in which the actin-binding sites on the FH2 domain are sterically occluded. However biochemical data suggest the full-length mDia1 is a dimer in solution (2 FH2 + 2 DID). Based on the crystal structure, we have generated possible dimer models and found that architectures of all of these models are incompatible with binding to actin filament but not to actin monomer. Furthermore, we show that the minimal functional monomeric unit in the FH2 domain, termed the bridge element, can be inhibited by isolated monomeric DID. NMR data on the bridge-DID system revealed that at least one of the two actin-binding sites on the bridge element is accessible to actin monomer in the inhibited state. Our findings suggest that autoinhibition in the native DRF dimer involves steric hindrance with the actin filament. Although the structure of a full-length DRF would be required for clarification of the presented models, our work here provides the first structural insights into the mechanism of the DRF autoinhibition.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, Texas, United States of America.