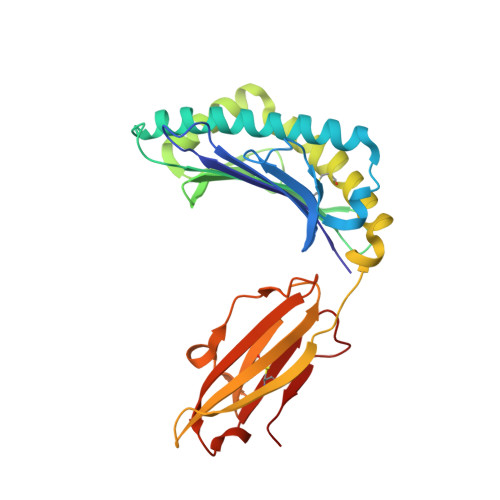
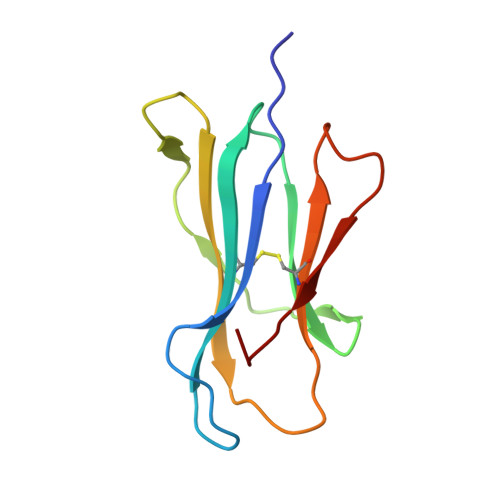
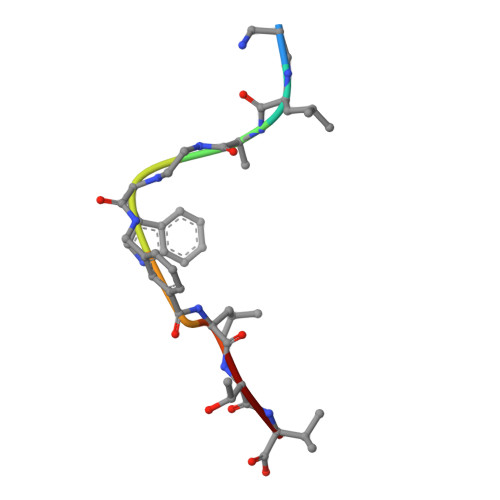
Crystal structures of HLA-A*0201 complexed with Melan-A/MART-1(26(27L)-35) peptidomimetics reveal conformational heterogeneity and highlight degeneracy of T cell recognition.
Douat-Casassus, C., Borbulevych, O., Tarbe, M., Gervois, N., Jotereau, F., Baker, B.M., Quideau, S.(2010) J Med Chem 53: 7061-7066
- PubMed: 20806940
- DOI: https://doi.org/10.1021/jm100683p
- Primary Citation of Related Structures:
3O3A, 3O3B, 3O3D, 3O3E - PubMed Abstract:
There is growing interest in using tumor associated antigens presented by class I major histocompatibility complex (MHC-I) proteins as cancer vaccines. As native peptides are poorly stable in biological fluids, researchers have sought to engineer synthetic peptidomimetics with greater biostability. Here, we demonstrate that antigenic peptidomimetics of the Melan-A/MART-1(26(27L)-35) melanoma antigen adopt strikingly different conformations when bound to MHC-I, highlighting the degeneracy of T cell recognition and revealing the challenges associated with mimicking native peptide conformation.
Organizational Affiliation:
Institut des Sciences Moléculaires (UMR-CNRS 5255) and Institut Européen de Chimie et Biologie (IECB), Université de Bordeaux, 2 Rue Robert Escarpit, 33607 Pessac, France.