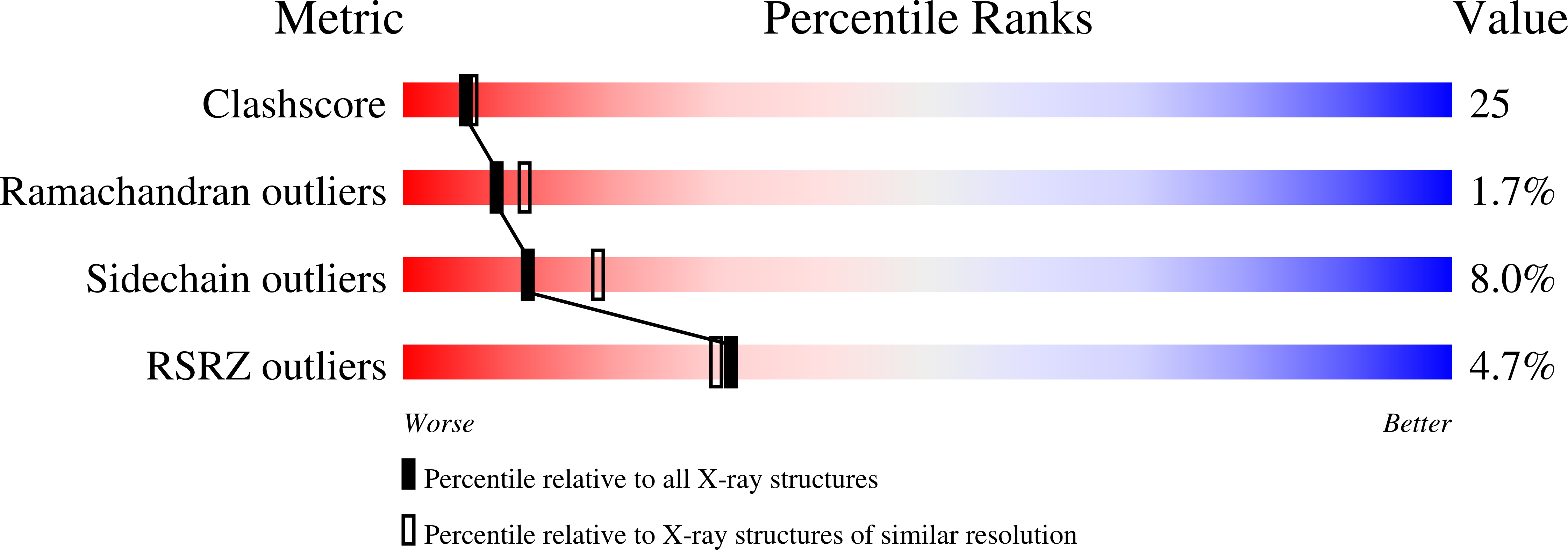Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Jimenez-Menendez, N., Fernandez-Millan, P., Rubio-Cosials, A., Arnan, C., Montoya, J., Jacobs, H.T., Bernado, P., Coll, M., Uson, I., Sola, M.(2010) Nat Struct Mol Biol 17: 891-893
- PubMed: 20543826
- DOI: https://doi.org/10.1038/nsmb.1859
- Primary Citation of Related Structures:
3N6S, 3N7Q - PubMed Abstract:
The regulation of mitochondrial DNA (mtDNA) processes is slowly being characterized at a structural level. We present here crystal structures of human mitochondrial regulator mTERF, a transcription termination factor also implicated in replication pausing, in complex with double-stranded DNA oligonucleotides containing the tRNA(Leu)(UUR) gene sequence. mTERF comprises nine left-handed helical tandem repeats that form a left-handed superhelix, the Zurdo domain.
Organizational Affiliation:
Institut de Biologia Molecular de Barcelona, Parc Cientific de Barcelona, Barcelona, Spain.