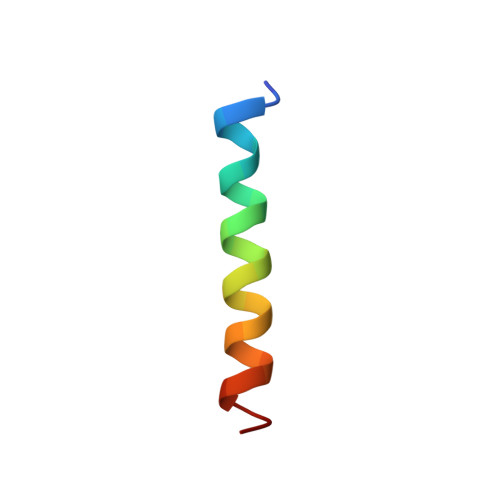Spatial structure of the M3 transmembrane segment of the nicotinic acetylcholine receptor alpha subunit.
Lugovskoy, A.A., Maslennikov, I.V., Utkin, Y.N., Tsetlin, V.I., Cohen, J.B., Arseniev, A.S.(1998) Eur J Biochem 255: 455-461
- PubMed: 9716388
- DOI: https://doi.org/10.1046/j.1432-1327.1998.2550455.x
- Primary Citation of Related Structures:
3MRA - PubMed Abstract:
The three-dimensional structure of a synthetic peptide corresponding to the putative transmembrane segment M3 (amino acid residues 277-301) of the alpha subunit of the nicotinic acetylcholine receptor from Torpedo californica has been studied by means of two-dimensional 1H-NMR spectroscopy in a chloroform/methanol (1:1) mixture containing 0.1 M LiClO4. Complete resonance assignment has been performed using double-quantum-filtered COSY (DQF-COSY), TOCSY and NOESY spectra. The spatial structure has been calculated using the Diana program on the basis of integrated intensities of NOESY spectra. HN-C(alpha)H and HC(alpha)-C(beta)H spin-spin coupling constants. Residues 279-297 of M3 form a right-handed helix (root mean square deviation is 0.032 nm for backbone atoms and 0.088 nm for all heavy atoms). The conformations of the 17 side chains have been unambiguously determined. The obtained structure is in accord with the photolabeling pattern of the membrane nicotinic acetylcholine receptor (nAChR) which suggests alpha-helical structure of M3 in the labeled portion [Blanton, M. P. & Cohen, J. B. (1994) Biochemistry 33, 2859-2872].
Organizational Affiliation:
Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow.