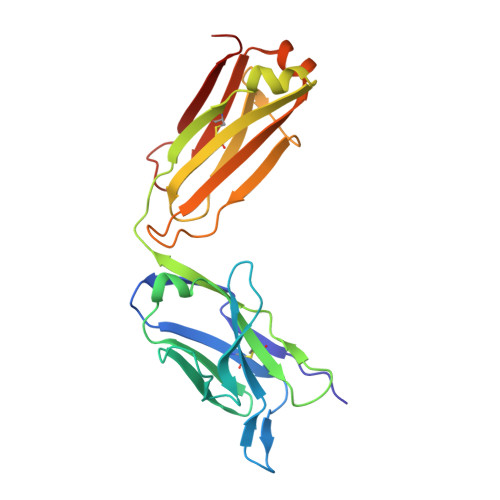
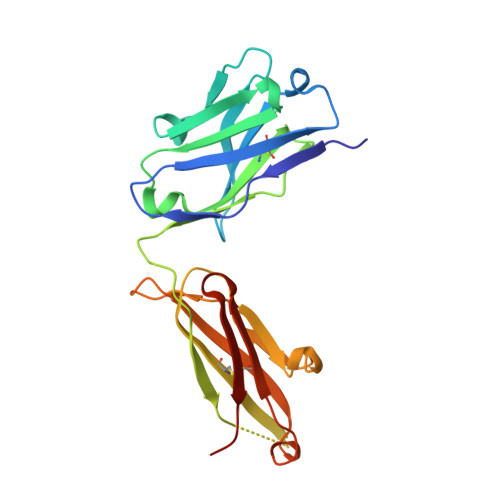
Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region.
Nicely, N.I., Dennison, S.M., Spicer, L., Scearce, R.M., Kelsoe, G., Ueda, Y., Chen, H., Liao, H.X., Alam, S.M., Haynes, B.F.(2010) Nat Struct Mol Biol 17: 1492-1494
- PubMed: 21076400
- DOI: https://doi.org/10.1038/nsmb.1944
- Primary Citation of Related Structures:
3MNV - PubMed Abstract:
The monoclonal antibody 13H11 shares part of its epitope in the HIV-1 gp41 membrane-proximal external region (MPER) with the rare, broadly neutralizing human antibody 2F5. Although 13H11 partially cross-blocked 2F5 binding, 13H11 is non-neutralizing and does not block 2F5 neutralization. We show that unlike 2F5, 13H11 binds to a well-defined helical MPER structure that is consistent with the structure of gp41 in a post-fusion six-helix bundle conformation.
Organizational Affiliation:
Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina, USA. nathan.nicely@duke.edu