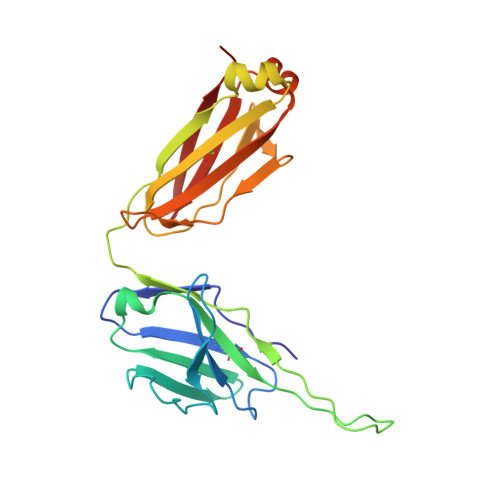
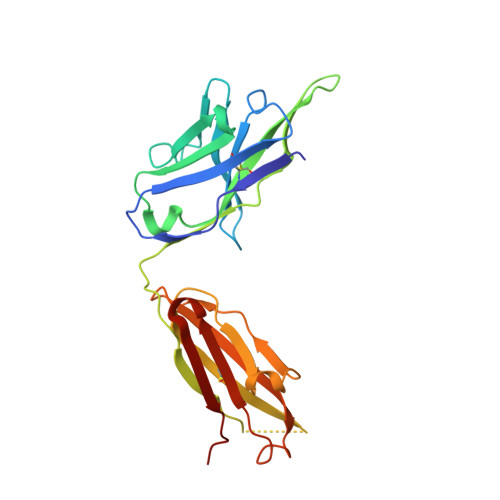
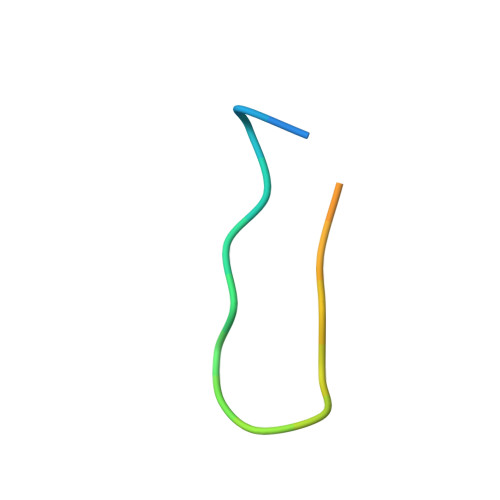
Conserved structural elements in the V3 crown of HIV-1 gp120.
Jiang, X., Burke, V., Totrov, M., Williams, C., Cardozo, T., Gorny, M.K., Zolla-Pazner, S., Kong, X.P.(2010) Nat Struct Mol Biol 17: 955-961
- PubMed: 20622876
- DOI: https://doi.org/10.1038/nsmb.1861
- Primary Citation of Related Structures:
3GO1, 3MLR, 3MLS, 3MLT, 3MLU, 3MLV, 3MLW, 3MLX, 3MLY, 3MLZ - PubMed Abstract:
Binding of the third variable region (V3) of the HIV-1 envelope glycoprotein gp120 to the cell-surface coreceptors CCR5 or CXCR4 during viral entry suggests that there are conserved structural elements in this sequence-variable region. These conserved elements could serve as epitopes to be targeted by a vaccine against HIV-1. Here we perform a systematic structural analysis of representative human anti-V3 monoclonal antibodies in complex with V3 peptides, revealing that the crown of V3 has four conserved structural elements: an arch, a band, a hydrophobic core and the peptide backbone. These are either unaffected by or are subject to minimal sequence variation. As these regions are targeted by cross-clade neutralizing human antibodies, they provide a blueprint for the design of vaccine immunogens that could elicit broadly cross-reactive protective antibodies.
Organizational Affiliation:
Department of Biochemistry, New York University School of Medicine, New York, New York, USA.