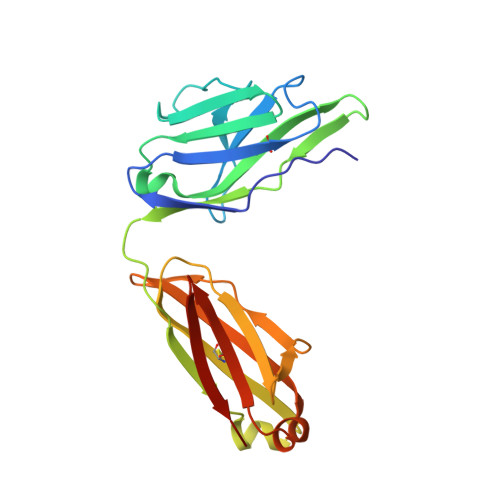
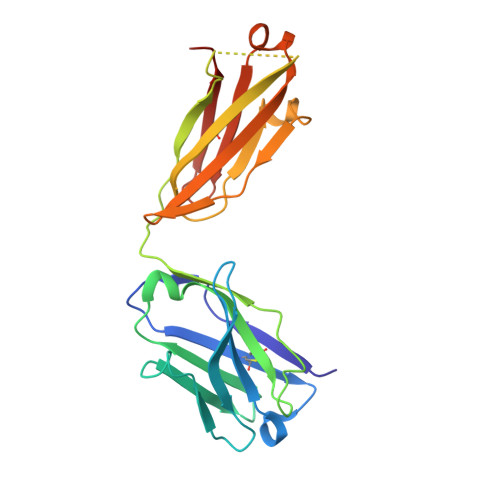
cDNA sequence and Fab crystal structure of HL4E10, a hamster IgG lambda light chain antibody stimulatory for gammadelta T cells.
Verdino, P., Witherden, D.A., Podshivalova, K., Rieder, S.E., Havran, W.L., Wilson, I.A.(2011) PLoS One 6: e19828-e19828
- PubMed: 21629689
- DOI: https://doi.org/10.1371/journal.pone.0019828
- Primary Citation of Related Structures:
3MJ8 - PubMed Abstract:
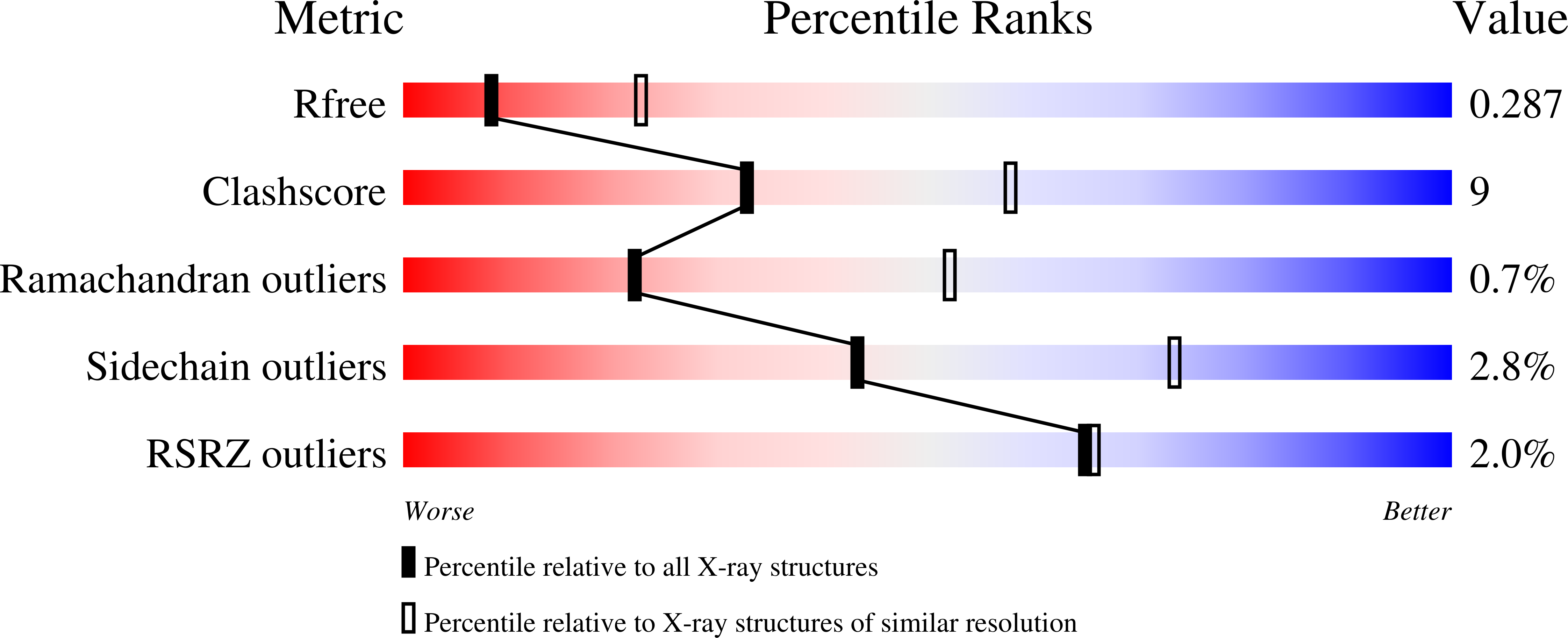
Hamsters are widely used to generate monoclonal antibodies against mouse, rat, and human antigens, but sequence and structural information for hamster immunoglobulins is sparse. To our knowledge, only three hamster IgG sequences have been published, all of which use kappa light chains, and no three-dimensional structure of a hamster antibody has been reported. We generated antibody HL4E10 as a probe to identify novel costimulatory molecules on the surface of γδ T cells which lack the traditional αβ T cell co-receptors CD4, CD8, and the costimulatory molecule CD28. HL4E10 binding to γδ T cell, surface-expressed, Junctional Adhesion Molecule-Like (JAML) protein leads to potent costimulation via activation of MAP kinase pathways and cytokine production, resulting in cell proliferation. The cDNA sequence of HL4E10 is the first example of a hamster lambda light chain and only the second known complete hamster heavy chain sequence. The crystal structure of the HL4E10 Fab at 2.95 Å resolution reveals a rigid combining site with pockets faceted by solvent-exposed tyrosine residues, which are structurally optimized for JAML binding. The characterization of HL4E10 thus comprises a valuable addition to the spartan database of hamster immunoglobulin genes and structures. As the HL4E10 antibody is uniquely costimulatory for γδ T cells, humanized versions thereof may be of clinical relevance in treating γδ T cell dysfunction-associated diseases, such as chronic non-healing wounds and cancer.
Organizational Affiliation:
Department of Molecular Biology, The Scripps Research Institute, La Jolla, California, United States of America.