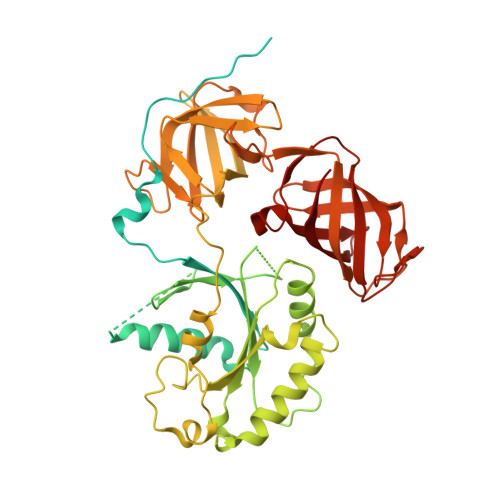
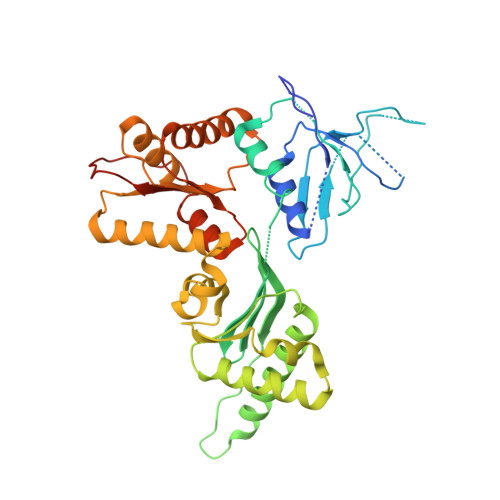
Structure of the Dom34-Hbs1 complex and implications for no-go decay
Chen, L., Muhlrad, D., Hauryliuk, V., Cheng, Z., Lim, M.K., Shyp, V., Parker, R., Song, H.(2010) Nat Struct Mol Biol 17: 1233-1240
- PubMed: 20890290
- DOI: https://doi.org/10.1038/nsmb.1922
- Primary Citation of Related Structures:
3MCA - PubMed Abstract:
No-go decay (NGD) targets mRNAs with stalls in translation elongation for endonucleolytic cleavage in a process involving the Dom34 and Hbs1 proteins. The crystal structure of a Schizosaccharomyces pombe Dom34-Hbs1 complex reveals an overall shape similar to that of eRF1-eRF3-GTP and EF-Tu-tRNA-GDPNP. Similarly to eRF1 and GTP binding to eRF3, Dom34 and GTP bind to Hbs1 with strong cooperativity, and Dom34 acts as a GTP-dissociation inhibitor (GDI). A marked conformational change in Dom34 occurs upon binding to Hbs1, leading Dom34 to resemble a portion of a tRNA and to position a conserved basic region in a position expected to be near the peptidyl transferase center. These results support the idea that the Dom34-Hbs1 complex functions to terminate translation and thereby commit mRNAs to NGD. Consistent with this role, NGD at runs of arginine codons, which cause a strong block to elongation, is independent of the Dom34-Hbs1 complex.
Organizational Affiliation:
Laboratory of Macromolecular Structure, Institute of Molecular and Cell Biology, Proteos, Singapore.