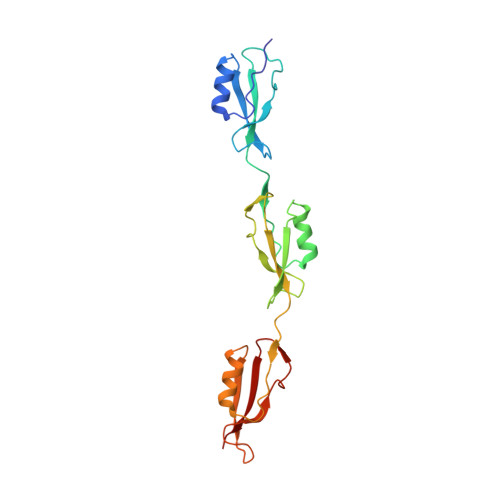
The Extended Conformation of the 2.9-A Crystal Structure of the Three-PASTA Domain of a Ser/Thr Kinase from the Human Pathogen Staphylococcus aureus
Paracuellos, P., Ballandras, A., Robert, X., Kahn, R., Herve, M., Mengin-Lecreulx, D., Cozzone, A.J., Duclos, B., Gouet, P.(2010) J Mol Biol 404: 847-858
- PubMed: 20965199
- DOI: https://doi.org/10.1016/j.jmb.2010.10.012
- Primary Citation of Related Structures:
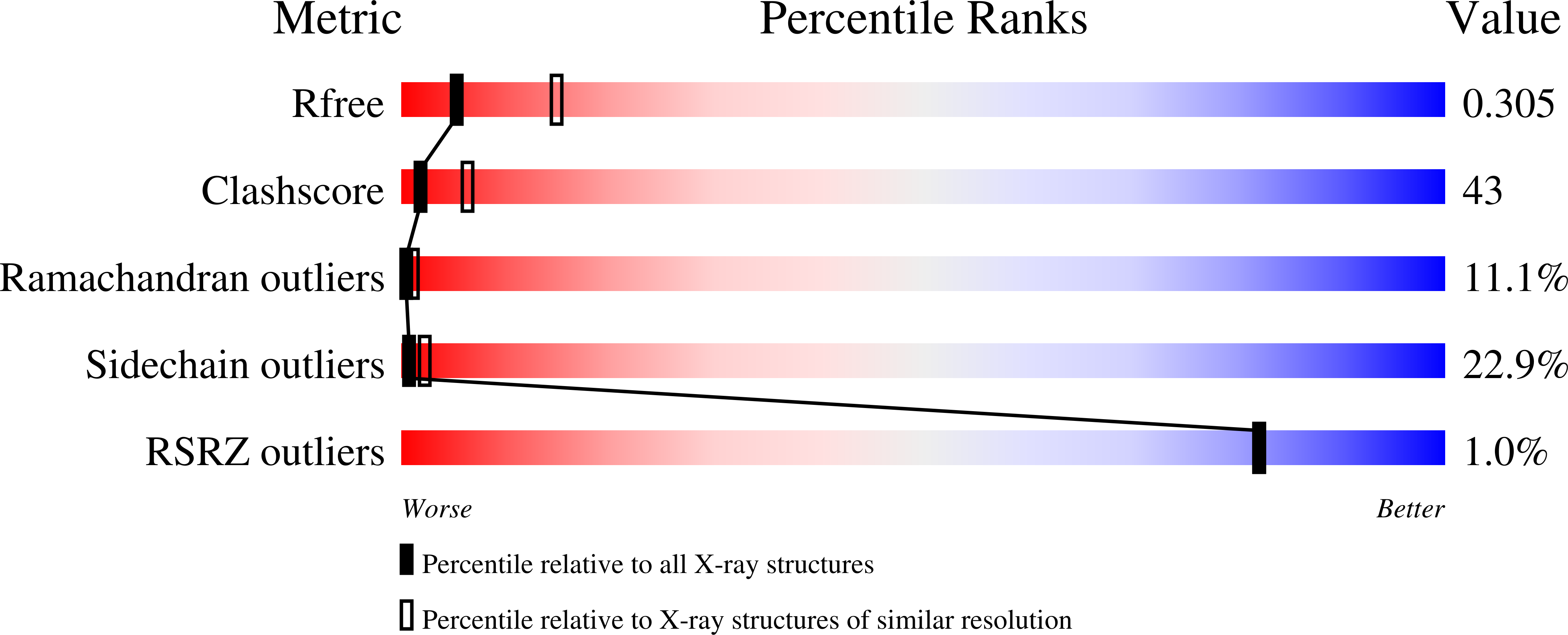
3M9G - PubMed Abstract:
PASTA (penicillin-binding protein and serine/threonine kinase associated) modules are found in penicillin-binding proteins and bacterial serine/threonine kinases mainly from Gram-positive Firmicutes and Actinobacteria. They may act as extracellular sensors by binding peptidoglycan fragments. We report here the first crystal structure of a multiple-PASTA domain from Ser/Thr kinase, that of the protein serine/threonine kinase 1 (Stk1) from the Firmicute Staphylococcus aureus. The extended conformation of the three PASTA subunits differs strongly from the compact conformation observed in the two-PASTA domain of penicillin-binding protein PBP2x, whereas linear conformations were also reported for two-subunit fragments of the four-PASTA domain of the Actinobacteria Mycobacterium tuberculosis studied by liquid NMR. Thus, a stretched organization appears to be the signature of modular PASTA domains in Ser/Thr kinases. Signal transduction to the kinase domain is supposed to occur via dimerization and ligand binding. A conserved X-shaped crystallographic dimer stabilized by intermolecular interactions between the second PASTA subunits of each monomer is observed in the two crystal forms of Stk1 that we managed to crystallize. Extracellular PASTA domains are composed of at least two subunits, and this molecular assembly is a plausible candidate for the biological dimer. We have also performed docking experiments, which predict that the hinge regions of the PASTA domain can accommodate peptidoglycan. Finally, a three-dimensional homology molecular model of full-length Stk1 was generated, suggesting an interaction between the kinase domain and the cytoplasmic face of the plasma membrane via a eukaryotic-like juxtamembrane domain. A comprehensive activation mechanism for bacterial Ser/Thr kinases is proposed with the support of these structural data.
Organizational Affiliation:
Laboratoire de Phosphorylation des Protéines et Pathogénie Bactérienne, Institut de Biologie et Chimie des Protéines, UMR 5086-CNRS/Université de Lyon, IFR128 BioSciences Gerland Lyon Sud, 7 Passage du Vercors, F-69367 Lyon cedex 07, France.